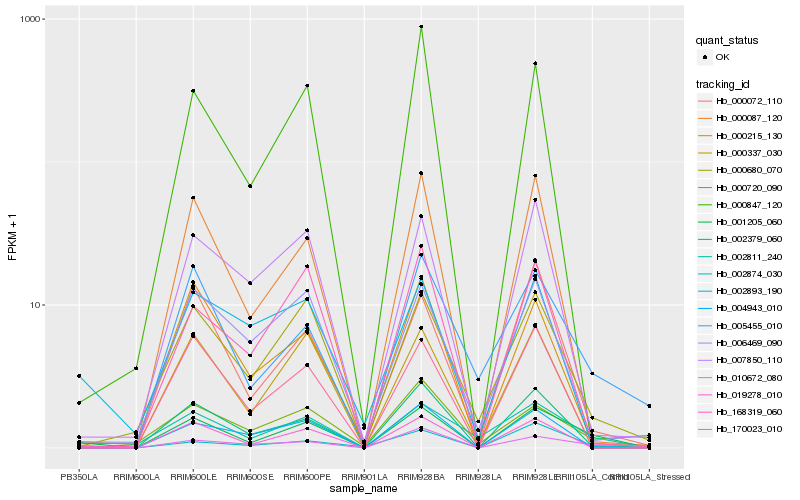
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_060 |
0.0 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 2 |
Hb_010672_080 |
0.2023475148 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 3 |
Hb_000680_070 |
0.2098324383 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002811_240 |
0.2106361356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002379_060 |
0.2121846471 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 6 |
Hb_019278_010 |
0.2233547059 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 7 |
Hb_005455_010 |
0.2253834755 |
- |
- |
PREDICTED: probable acyl-activating enzyme 18, peroxisomal isoform X1 [Jatropha curcas] |
| 8 |
Hb_000087_120 |
0.2257024121 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 9 |
Hb_000072_110 |
0.2282593962 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 10 |
Hb_002874_030 |
0.2328713895 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 11 |
Hb_000337_030 |
0.2342857042 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_170023_010 |
0.2348363184 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 13 |
Hb_007850_110 |
0.2354830607 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 14 |
Hb_000215_130 |
0.2360778184 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 15 |
Hb_000720_090 |
0.2401839534 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_004943_010 |
0.2423177683 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_168319_060 |
0.2428730866 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 18 |
Hb_002893_190 |
0.2440060845 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000847_120 |
0.2443884496 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 20 |
Hb_006469_090 |
0.2449811253 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |