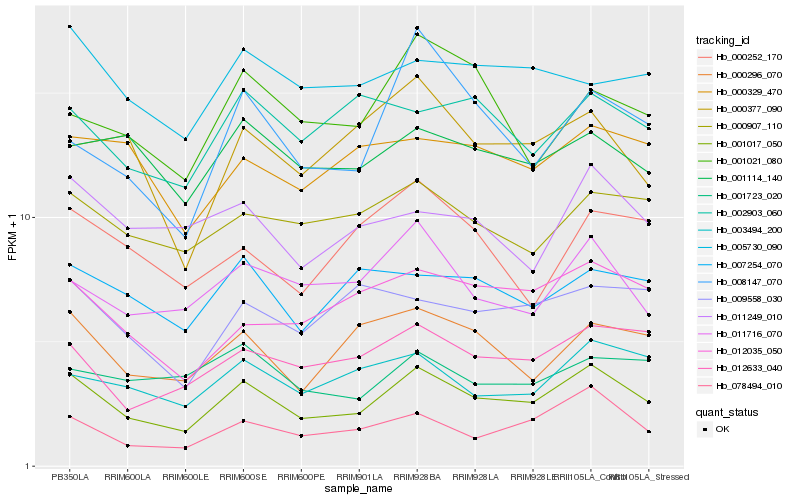
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001017_050 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_078494_010 |
0.1037967478 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like [Jatropha curcas] |
| 3 |
Hb_012633_040 |
0.108530894 |
- |
- |
- |
| 4 |
Hb_000296_070 |
0.1130119866 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 5 |
Hb_011716_070 |
0.1134534286 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012035_050 |
0.114304937 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 7 |
Hb_003494_200 |
0.1156329551 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 8 |
Hb_000907_110 |
0.1178865139 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 9 |
Hb_007254_070 |
0.1220734865 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_011249_010 |
0.1239152316 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 35, NatC auxiliary subunit isoform X1 [Jatropha curcas] |
| 11 |
Hb_008147_070 |
0.1240178034 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 12 |
Hb_001114_140 |
0.1248921197 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 13 |
Hb_000329_470 |
0.1262240589 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 14 |
Hb_005730_090 |
0.1263827167 |
- |
- |
PREDICTED: uncharacterized protein LOC105631484 [Jatropha curcas] |
| 15 |
Hb_009558_030 |
0.1264702985 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 16 |
Hb_002903_060 |
0.1273898246 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 17 |
Hb_001723_020 |
0.1291332648 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 18 |
Hb_001021_080 |
0.1292738844 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 19 |
Hb_000377_090 |
0.1294402616 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000252_170 |
0.1304060079 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |