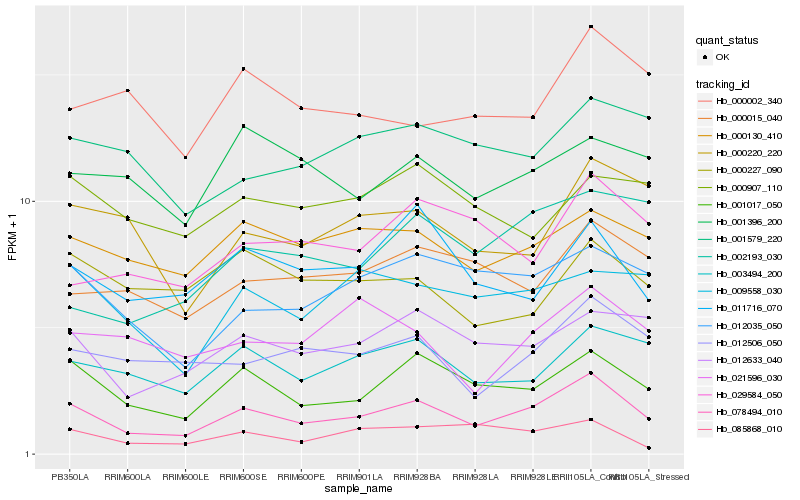
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078494_010 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like [Jatropha curcas] |
| 2 |
Hb_001017_050 |
0.1037967478 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_012633_040 |
0.1301497481 |
- |
- |
- |
| 4 |
Hb_003494_200 |
0.132572495 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 5 |
Hb_012035_050 |
0.1328267953 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 6 |
Hb_012506_050 |
0.1378525871 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011716_070 |
0.1409915028 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000130_410 |
0.1428293296 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 9 |
Hb_000220_220 |
0.1467884455 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 10 |
Hb_009558_030 |
0.1473985648 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 11 |
Hb_000015_040 |
0.1491371405 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001579_220 |
0.154029031 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 13 |
Hb_029584_050 |
0.1568867802 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3 [Jatropha curcas] |
| 14 |
Hb_001396_200 |
0.1579292681 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_021596_030 |
0.1585166704 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000002_340 |
0.1592865186 |
- |
- |
PREDICTED: peroxisomal bifunctional enzyme-like [Jatropha curcas] |
| 17 |
Hb_000227_090 |
0.1613011553 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 catalytic subunit [Jatropha curcas] |
| 18 |
Hb_002193_030 |
0.1624507378 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 19 |
Hb_085868_010 |
0.1632481174 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_000907_110 |
0.1634632045 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |