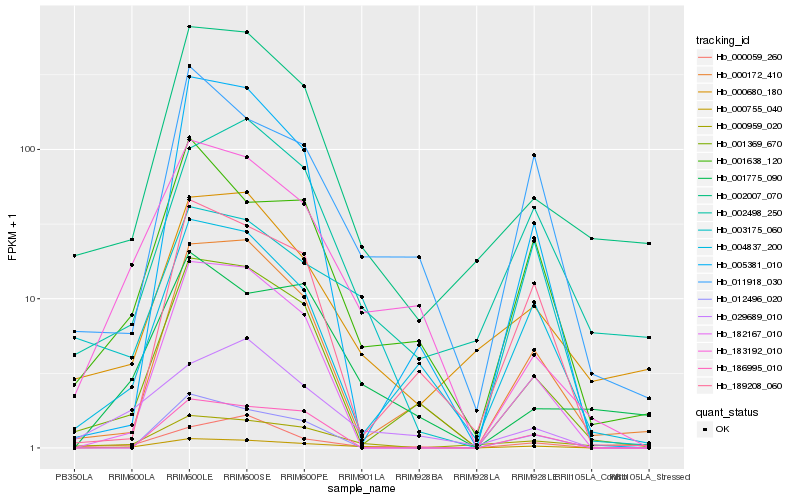
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000755_040 |
0.0 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 2 |
Hb_182167_010 |
0.1927211941 |
- |
- |
hypothetical protein PHAVU_001G0486001g, partial [Phaseolus vulgaris] |
| 3 |
Hb_183192_010 |
0.1988157293 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 4 |
Hb_001638_120 |
0.204351427 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 5 |
Hb_001369_670 |
0.2090757412 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_029689_010 |
0.2125092912 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_000059_260 |
0.2130944312 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: peroxidase 60 [Jatropha curcas] |
| 8 |
Hb_011918_030 |
0.2143990996 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 9 |
Hb_003175_060 |
0.214404248 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 10 |
Hb_012496_020 |
0.2159856116 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 11 |
Hb_002007_070 |
0.2159858734 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 12 |
Hb_004837_200 |
0.2163398817 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 13 |
Hb_000959_020 |
0.2164495171 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_001775_090 |
0.2167665711 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 15 |
Hb_000172_410 |
0.219021171 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_002498_250 |
0.2199091354 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000680_180 |
0.2203805081 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 18 |
Hb_186995_010 |
0.2215803119 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |
| 19 |
Hb_005381_010 |
0.2250824128 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 20 |
Hb_189208_060 |
0.2272163744 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |