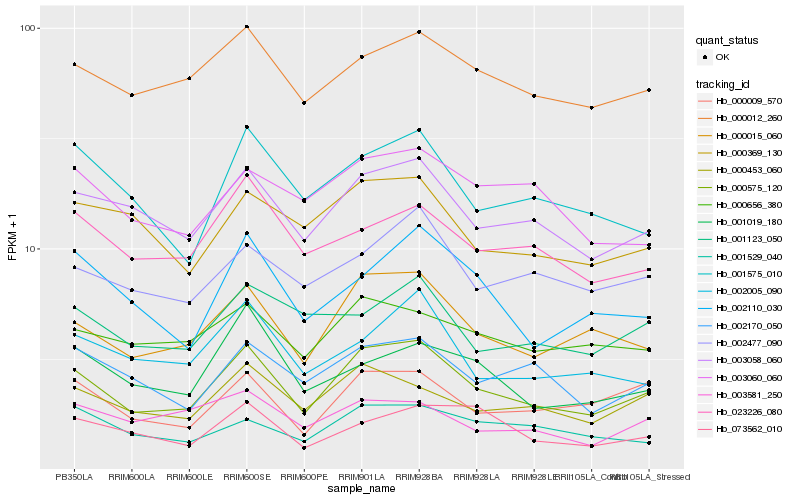
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000575_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s02060g [Populus trichocarpa] |
| 2 |
Hb_000015_060 |
0.0771742122 |
- |
- |
PREDICTED: uncharacterized protein LOC104239844 isoform X2 [Nicotiana sylvestris] |
| 3 |
Hb_003058_060 |
0.0923542703 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 4 |
Hb_000453_060 |
0.0981605096 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g11690 [Jatropha curcas] |
| 5 |
Hb_000009_570 |
0.1040120035 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Populus euphratica] |
| 6 |
Hb_000012_260 |
0.108006569 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 7 |
Hb_002110_030 |
0.1080294601 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_002170_050 |
0.1103602714 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |
| 9 |
Hb_001575_010 |
0.1107917248 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002005_090 |
0.1114608148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000369_130 |
0.1144754798 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 12 |
Hb_000656_380 |
0.1153815911 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 13 |
Hb_002477_090 |
0.1155151417 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 14 |
Hb_023226_080 |
0.1158773478 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001529_040 |
0.1168049191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 16 |
Hb_001019_180 |
0.1197417011 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 17 |
Hb_001123_050 |
0.121221989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003060_060 |
0.1222789561 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 19 |
Hb_073562_010 |
0.1223906233 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 20 |
Hb_003581_250 |
0.1226907068 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |