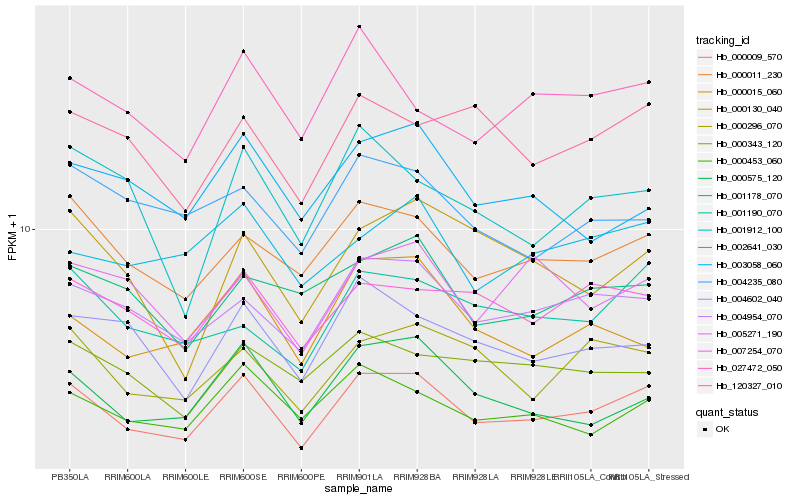
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_570 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Populus euphratica] |
| 2 |
Hb_000011_230 |
0.0967323625 |
- |
- |
PREDICTED: bifunctional protein FolD 4, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_004954_070 |
0.0988721538 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 4 |
Hb_000575_120 |
0.1040120035 |
- |
- |
hypothetical protein POPTR_0011s02060g [Populus trichocarpa] |
| 5 |
Hb_000453_060 |
0.1053868341 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g11690 [Jatropha curcas] |
| 6 |
Hb_000296_070 |
0.106366737 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 7 |
Hb_004602_040 |
0.1125497084 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_007254_070 |
0.1138433143 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001190_070 |
0.1153440234 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 10 |
Hb_000015_060 |
0.1154288625 |
- |
- |
PREDICTED: uncharacterized protein LOC104239844 isoform X2 [Nicotiana sylvestris] |
| 11 |
Hb_001912_100 |
0.1178335718 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 12 |
Hb_027472_050 |
0.1178525182 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 13 |
Hb_120327_010 |
0.118836886 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 14 |
Hb_005271_190 |
0.1194590687 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000130_040 |
0.1207036289 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 16 |
Hb_002641_030 |
0.1213774728 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 17 |
Hb_001178_070 |
0.122518746 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 18 |
Hb_000343_120 |
0.1235153726 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 19 |
Hb_003058_060 |
0.1236866817 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 20 |
Hb_004235_080 |
0.1245152735 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |