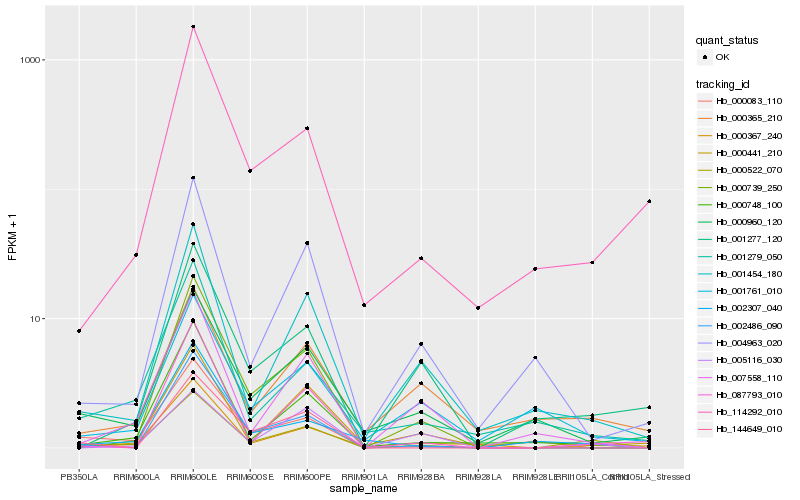
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000522_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 2 |
Hb_000367_240 |
0.194674797 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 3 |
Hb_001454_180 |
0.2038624138 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 4 |
Hb_000748_100 |
0.2063420966 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 5 |
Hb_007558_110 |
0.2130762339 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 6 |
Hb_000739_250 |
0.2174762882 |
- |
- |
PREDICTED: uncharacterized protein LOC105120821 [Populus euphratica] |
| 7 |
Hb_000083_110 |
0.2203506787 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 8 |
Hb_000960_120 |
0.2211190431 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 9 |
Hb_001279_050 |
0.2223000898 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 10 |
Hb_002486_090 |
0.226385185 |
- |
- |
hypothetical protein POPTR_0005s06910g [Populus trichocarpa] |
| 11 |
Hb_001277_120 |
0.2275641329 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 12 |
Hb_000365_210 |
0.2287800044 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 13 |
Hb_004963_020 |
0.2296983461 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 14 |
Hb_087793_010 |
0.2301768043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_114292_010 |
0.2307157456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002307_040 |
0.2322525538 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_144649_010 |
0.2339449541 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 18 |
Hb_000441_210 |
0.2343283626 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 19 |
Hb_001761_010 |
0.2355603712 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 20 |
Hb_005116_030 |
0.2369799855 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |