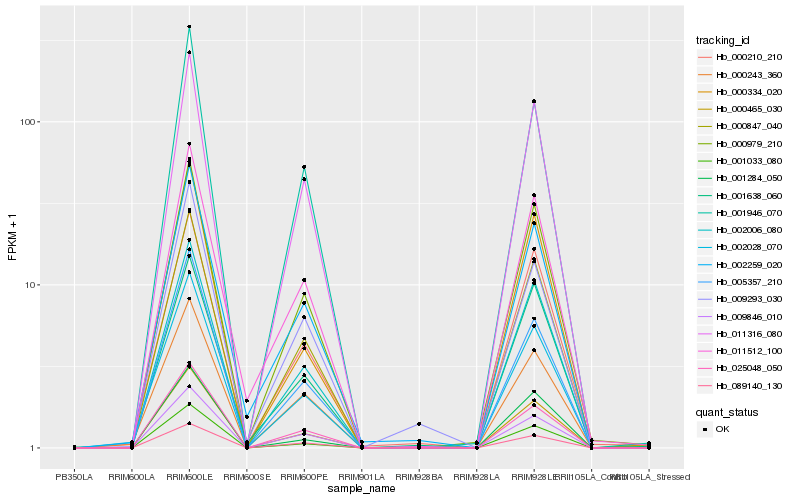
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_030 |
0.0 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_009846_010 |
0.0995450623 |
- |
- |
PREDICTED: uncharacterized protein LOC105643801 [Jatropha curcas] |
| 3 |
Hb_000210_210 |
0.1009776646 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000243_360 |
0.1025212559 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 5 |
Hb_000847_040 |
0.1029520803 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 6 |
Hb_001033_080 |
0.1061692839 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 7 |
Hb_002006_080 |
0.1062126565 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Citrus sinensis] |
| 8 |
Hb_000979_210 |
0.1063536938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_025048_050 |
0.1108089779 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 10 |
Hb_002028_070 |
0.1114768014 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 11 |
Hb_011316_080 |
0.1121032779 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 12 |
Hb_005357_210 |
0.1124366831 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein MERISTEM L1 [Jatropha curcas] |
| 13 |
Hb_002259_020 |
0.113328901 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 14 |
Hb_001284_050 |
0.1135159026 |
- |
- |
PREDICTED: uncharacterized protein LOC100781779 isoform X1 [Glycine max] |
| 15 |
Hb_001946_070 |
0.1143865363 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_001638_060 |
0.1148040291 |
- |
- |
PREDICTED: rho GTPase-activating protein 3-like isoform X1 [Populus euphratica] |
| 17 |
Hb_009293_030 |
0.1156813289 |
- |
- |
PREDICTED: salicylate carboxymethyltransferase [Jatropha curcas] |
| 18 |
Hb_089140_130 |
0.1158625138 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 19 |
Hb_000334_020 |
0.1165350451 |
- |
- |
PREDICTED: dynein light chain 2, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_011512_100 |
0.1168105751 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X1 [Jatropha curcas] |