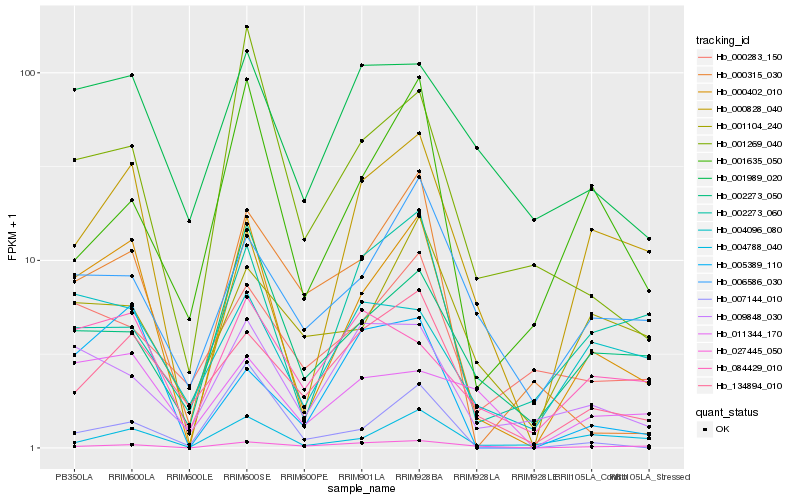
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_010 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |
| 2 |
Hb_004788_040 |
0.2324232545 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_005389_110 |
0.2413545773 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 4 |
Hb_001269_040 |
0.2426012052 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_002273_050 |
0.2427689517 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_134894_010 |
0.2481892647 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 7 |
Hb_009848_030 |
0.2507570223 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 8 |
Hb_001635_050 |
0.2570472561 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Nicotiana sylvestris] |
| 9 |
Hb_000315_030 |
0.258555539 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 10 |
Hb_006586_030 |
0.2588128386 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 11 |
Hb_007144_010 |
0.2596115628 |
- |
- |
- |
| 12 |
Hb_001989_020 |
0.2606837511 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 13 |
Hb_000828_040 |
0.2609263616 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 14 |
Hb_084429_010 |
0.2633612606 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Populus euphratica] |
| 15 |
Hb_004096_080 |
0.2634385882 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 16 |
Hb_027445_050 |
0.2666798668 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 17 |
Hb_002273_060 |
0.2674068035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000283_150 |
0.2680842159 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 19 |
Hb_001104_240 |
0.268460963 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_011344_170 |
0.2695478427 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 4-like, partial [Camelina sativa] |