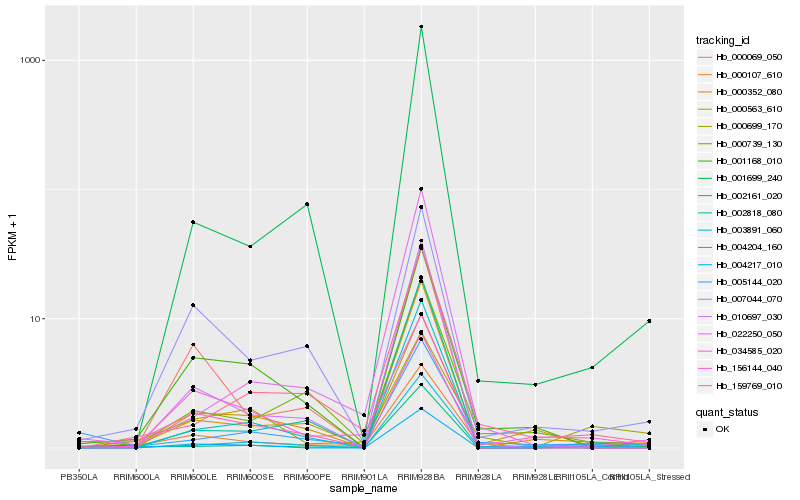
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_610 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_159769_010 |
0.1628885445 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 3 |
Hb_000352_080 |
0.1769645245 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 4 |
Hb_003891_060 |
0.1834434017 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 5 |
Hb_034585_020 |
0.1837137964 |
- |
- |
- |
| 6 |
Hb_004217_010 |
0.1851180622 |
- |
- |
putative wall-associated kinase family protein [Populus trichocarpa] |
| 7 |
Hb_000563_610 |
0.186912857 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 8 |
Hb_007044_070 |
0.1870950296 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 9 |
Hb_005144_020 |
0.1873839447 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 10 |
Hb_000739_130 |
0.1880509695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010697_030 |
0.1889921781 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 12 |
Hb_001699_240 |
0.1901321662 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 13 |
Hb_002161_020 |
0.190440175 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 14 |
Hb_004204_160 |
0.1908087147 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 15 |
Hb_000069_050 |
0.1915045152 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 16 |
Hb_001168_010 |
0.1916537311 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 17 |
Hb_022250_050 |
0.1918187094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_156144_040 |
0.1920248224 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000699_170 |
0.1920401315 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 20 |
Hb_002818_080 |
0.1957694989 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |