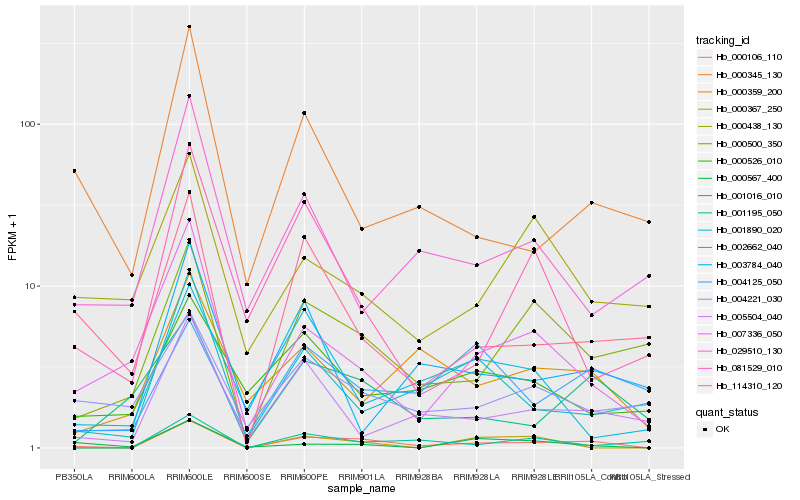
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_110 |
0.0 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 2 |
Hb_000500_350 |
0.2644498397 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 3 |
Hb_007336_050 |
0.2672723378 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 4 |
Hb_001016_010 |
0.2749360611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005504_040 |
0.2753352107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_114310_120 |
0.2816711533 |
- |
- |
PREDICTED: 50S ribosomal protein L31, chloroplastic [Eucalyptus grandis] |
| 7 |
Hb_000359_200 |
0.2840418054 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 8 |
Hb_001890_020 |
0.2848379213 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 9 |
Hb_004221_030 |
0.2906293499 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001195_050 |
0.2947046612 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 11 |
Hb_000526_010 |
0.3031393544 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 12 |
Hb_000345_130 |
0.3037602476 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_004125_050 |
0.3048774121 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 14 |
Hb_000567_400 |
0.3073704618 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 15 |
Hb_003784_040 |
0.3089500899 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000438_130 |
0.310826581 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 17 |
Hb_081529_010 |
0.312765425 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000367_250 |
0.3135684949 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_029510_130 |
0.3141728783 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 20 |
Hb_002662_040 |
0.3146437584 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |