| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
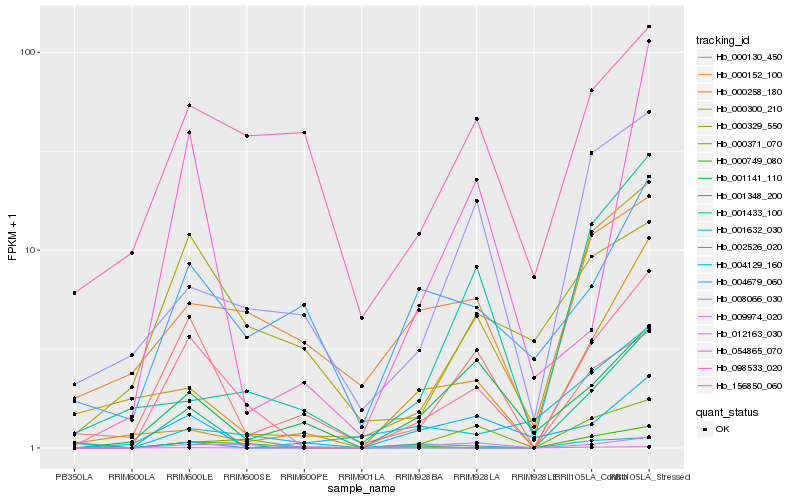
Hb_156850_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635248 [Jatropha curcas] |
| 2 |
Hb_000371_070 |
0.2425447236 |
- |
- |
PREDICTED: uncharacterized protein LOC103863319 [Brassica rapa] |
| 3 |
Hb_009974_020 |
0.2604749152 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 4 |
Hb_001632_030 |
0.2885489682 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 5 |
Hb_008066_030 |
0.2894568865 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 6 |
Hb_001348_200 |
0.2952790432 |
- |
- |
- |
| 7 |
Hb_004129_160 |
0.2972894404 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 8 |
Hb_054865_070 |
0.2988044657 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 9 |
Hb_001141_110 |
0.3011029485 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 10 |
Hb_000152_100 |
0.3081919935 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000329_550 |
0.3084027524 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 12 |
Hb_004679_060 |
0.3110640603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001433_100 |
0.3122894203 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_098533_020 |
0.3162994283 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 15 |
Hb_012163_030 |
0.3171412336 |
- |
- |
PREDICTED: uncharacterized protein LOC105647861 [Jatropha curcas] |
| 16 |
Hb_000749_080 |
0.3183555041 |
- |
- |
Alpha-expansin 12 precursor, putative [Ricinus communis] |
| 17 |
Hb_000300_210 |
0.3198876975 |
- |
- |
gd2b, putative [Ricinus communis] |
| 18 |
Hb_000130_450 |
0.3218841362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002526_020 |
0.3243780241 |
- |
- |
- |
| 20 |
Hb_000258_180 |
0.3257119944 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |