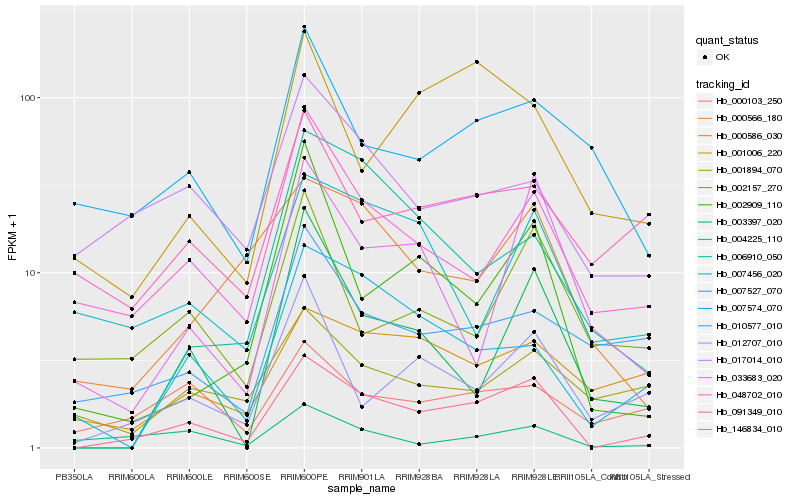
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146834_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_035385 [Theobroma cacao] |
| 2 |
Hb_048702_010 |
0.2318158786 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_012707_010 |
0.2403514599 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 4 |
Hb_007527_070 |
0.2439164616 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 5 |
Hb_000566_180 |
0.2513491014 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 6 |
Hb_003397_020 |
0.2538204453 |
- |
- |
- |
| 7 |
Hb_000103_250 |
0.2564771369 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 8 |
Hb_017014_010 |
0.2566607583 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000586_030 |
0.2579455285 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 10 |
Hb_010577_010 |
0.2583761215 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 11 |
Hb_007456_020 |
0.2590933002 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_033683_020 |
0.260406157 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 13 |
Hb_001006_220 |
0.2641952821 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 14 |
Hb_002909_110 |
0.2642618005 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 15 |
Hb_091349_010 |
0.2643636332 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 16 |
Hb_004225_110 |
0.2645512749 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 17 |
Hb_002157_270 |
0.2658678336 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 18 |
Hb_001894_070 |
0.2675592339 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007574_070 |
0.2678236777 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 20 |
Hb_006910_050 |
0.2681787558 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |