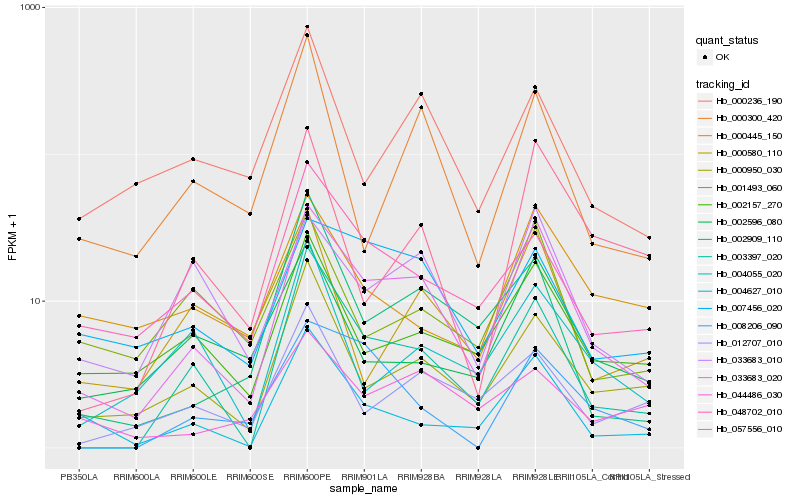
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033683_020 |
0.0 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 2 |
Hb_003397_020 |
0.175591993 |
- |
- |
- |
| 3 |
Hb_033683_010 |
0.1852927705 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 4 |
Hb_000950_030 |
0.1937764142 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 5 |
Hb_004627_010 |
0.1961085896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002157_270 |
0.201125439 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 7 |
Hb_057556_010 |
0.212069425 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 8 |
Hb_048702_010 |
0.2122982526 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_007456_020 |
0.2138968908 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_002596_080 |
0.2172982648 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 11 |
Hb_002909_110 |
0.2224882444 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 12 |
Hb_001493_060 |
0.2224940439 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 13 |
Hb_000580_110 |
0.2238805821 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 14 |
Hb_012707_010 |
0.2259472574 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 15 |
Hb_000236_190 |
0.2265062096 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 16 |
Hb_000300_420 |
0.226964259 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_008206_090 |
0.231093099 |
- |
- |
PREDICTED: uncharacterized protein LOC100811033 [Glycine max] |
| 18 |
Hb_000445_150 |
0.2318962238 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 19 |
Hb_044486_030 |
0.233612389 |
- |
- |
DMI1, partial [Cycas revoluta] |
| 20 |
Hb_004055_020 |
0.2337809038 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |