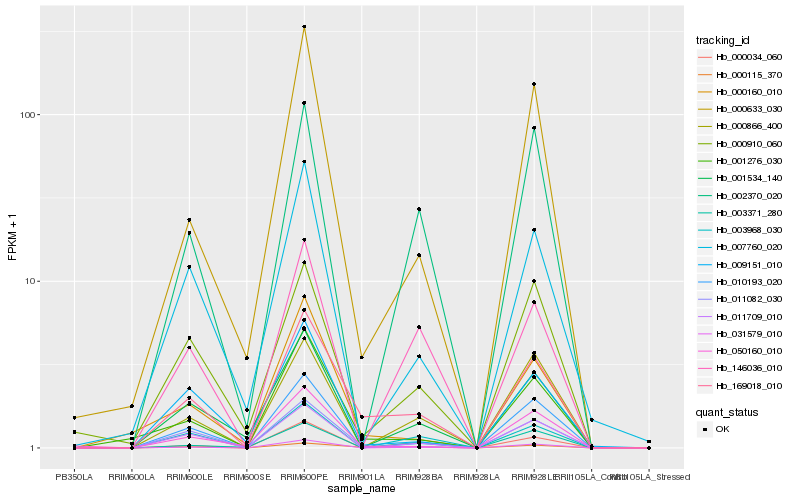
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169018_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s00800g [Populus trichocarpa] |
| 2 |
Hb_000115_370 |
0.087955593 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001276_030 |
0.1643961229 |
- |
- |
PREDICTED: uncharacterized protein LOC104881333 [Vitis vinifera] |
| 4 |
Hb_011082_030 |
0.1667538675 |
- |
- |
hypothetical protein RCOM_0852210 [Ricinus communis] |
| 5 |
Hb_031579_010 |
0.1762435738 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 6 |
Hb_011709_010 |
0.1762924982 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 7 |
Hb_003968_030 |
0.1845591817 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_010193_020 |
0.187199055 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 9 |
Hb_000160_010 |
0.1878037868 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 10 |
Hb_000633_030 |
0.1887337519 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 11 |
Hb_000910_060 |
0.1913628196 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 12 |
Hb_001534_140 |
0.1914116043 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 13 |
Hb_003371_280 |
0.1934082225 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_007760_020 |
0.195871102 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 15 |
Hb_000866_400 |
0.1971959953 |
- |
- |
- |
| 16 |
Hb_146036_010 |
0.1994862982 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 17 |
Hb_000034_060 |
0.1999429119 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 18 |
Hb_002370_020 |
0.2007906046 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 19 |
Hb_050160_010 |
0.2013935774 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 20 |
Hb_009151_010 |
0.2025856227 |
- |
- |
Peroxidase 3 [Morus notabilis] |