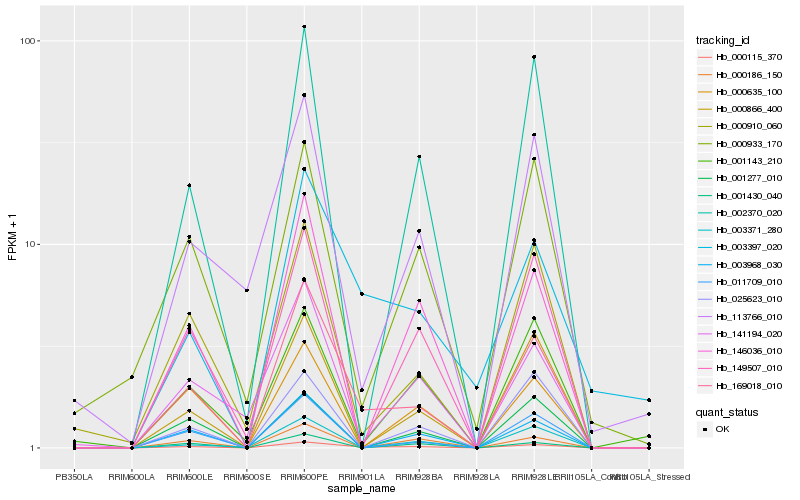
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_370 |
0.0 |
- |
- |
PREDICTED: nicotinamide adenine dinucleotide transporter 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_169018_010 |
0.087955593 |
- |
- |
hypothetical protein POPTR_0014s00800g [Populus trichocarpa] |
| 3 |
Hb_003968_030 |
0.1950774785 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_003397_020 |
0.196721909 |
- |
- |
- |
| 5 |
Hb_149507_010 |
0.1977350052 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 6 |
Hb_002370_020 |
0.1986828605 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 7 |
Hb_146036_010 |
0.2014325215 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 8 |
Hb_003371_280 |
0.2018465639 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000910_060 |
0.2064601176 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_000635_100 |
0.2100604054 |
- |
- |
- |
| 11 |
Hb_000866_400 |
0.2106204591 |
- |
- |
- |
| 12 |
Hb_000186_150 |
0.211392662 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 13 |
Hb_000933_170 |
0.2115845851 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 14 |
Hb_011709_010 |
0.2130194682 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 15 |
Hb_001430_040 |
0.2149752254 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 16 |
Hb_141194_020 |
0.2170737274 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 17 |
Hb_001143_210 |
0.2176945602 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 18 |
Hb_001277_010 |
0.2178981266 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_025623_010 |
0.2207173716 |
- |
- |
- |
| 20 |
Hb_113766_010 |
0.2208421434 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |