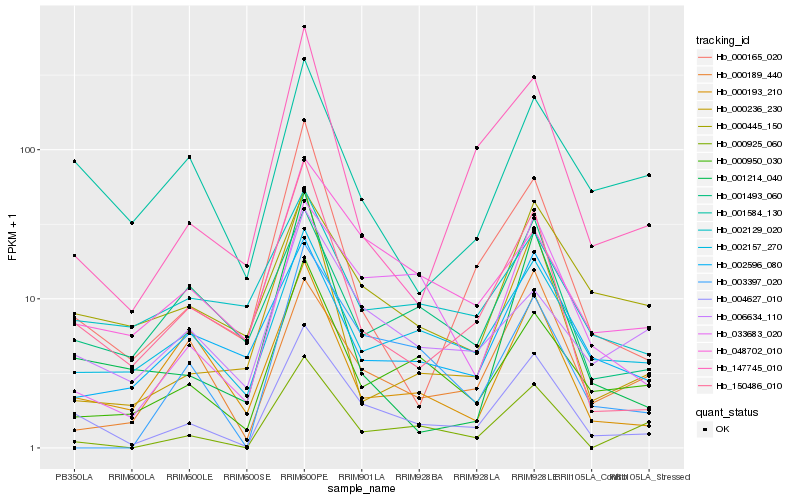
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004627_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_150486_010 |
0.1823595795 |
- |
- |
hypothetical protein CICLE_v100116733mg, partial [Citrus clementina] |
| 3 |
Hb_002157_270 |
0.1902910193 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 4 |
Hb_000950_030 |
0.1916229643 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 5 |
Hb_033683_020 |
0.1961085896 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 6 |
Hb_000925_060 |
0.1994408063 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 7 |
Hb_048702_010 |
0.2018165684 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000165_020 |
0.2027651985 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 9 |
Hb_003397_020 |
0.2102248603 |
- |
- |
- |
| 10 |
Hb_006634_110 |
0.2109271945 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 11 |
Hb_000445_150 |
0.2151313053 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 12 |
Hb_147745_010 |
0.2159649436 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 13 |
Hb_000193_210 |
0.2160308375 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 14 |
Hb_001493_060 |
0.2164708798 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 15 |
Hb_001214_040 |
0.2186888781 |
- |
- |
PREDICTED: protein trichome birefringence-like 3 [Jatropha curcas] |
| 16 |
Hb_001584_130 |
0.2205132921 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 17 |
Hb_002596_080 |
0.221901216 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 18 |
Hb_000236_230 |
0.2243661661 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000189_440 |
0.2272960645 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |
| 20 |
Hb_002129_020 |
0.2315382819 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |