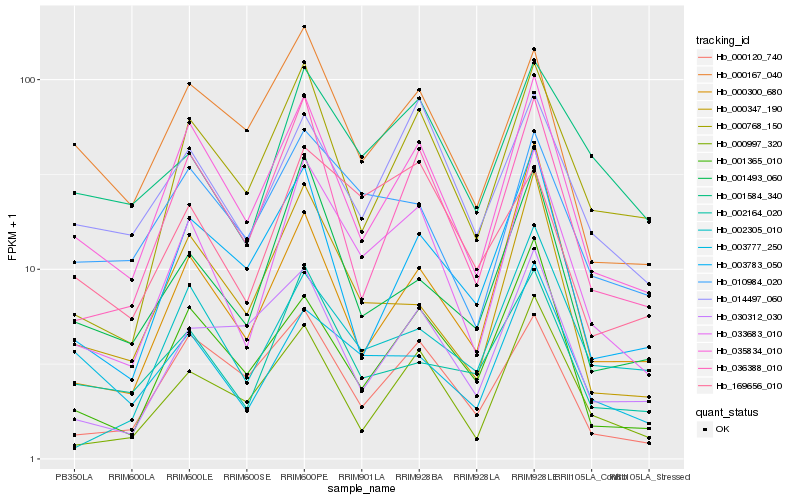
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033683_010 |
0.0 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 2 |
Hb_035834_010 |
0.1220985466 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_036388_010 |
0.1305459941 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 4 |
Hb_000768_150 |
0.1459079835 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_000997_320 |
0.1478467451 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 6 |
Hb_000347_190 |
0.1561163766 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 7 |
Hb_001365_010 |
0.161074539 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 8 |
Hb_001493_060 |
0.1614770839 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 9 |
Hb_000120_740 |
0.1635992369 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010984_020 |
0.1672655829 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 11 |
Hb_002164_020 |
0.1676929728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001584_340 |
0.1680224914 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 13 |
Hb_014497_060 |
0.1700739579 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 14 |
Hb_169656_010 |
0.1742754142 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_003783_050 |
0.1752615798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002305_010 |
0.1765273357 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000167_040 |
0.1815652362 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 18 |
Hb_030312_030 |
0.181659204 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000300_680 |
0.1822042435 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003777_250 |
0.1822347153 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |