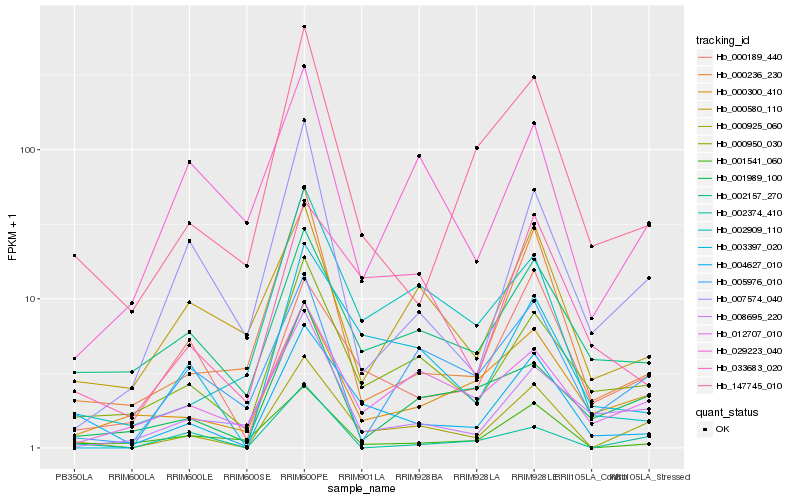
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000925_060 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 2 |
Hb_004627_010 |
0.1994408063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000950_030 |
0.202072577 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 4 |
Hb_003397_020 |
0.2197540847 |
- |
- |
- |
| 5 |
Hb_012707_010 |
0.227745121 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 6 |
Hb_000236_230 |
0.2305470977 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 7 |
Hb_005976_010 |
0.2411078634 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000300_410 |
0.2428733568 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 9 |
Hb_001541_060 |
0.244377563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002157_270 |
0.2461430059 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 11 |
Hb_001989_100 |
0.2467732981 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 12 |
Hb_033683_020 |
0.2504352164 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 13 |
Hb_007574_040 |
0.2505135047 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 14 |
Hb_000189_440 |
0.2508323967 |
- |
- |
hypothetical protein JCGZ_23470 [Jatropha curcas] |
| 15 |
Hb_008695_220 |
0.2512162042 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 16 |
Hb_147745_010 |
0.251299005 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 17 |
Hb_002374_410 |
0.2524466108 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_002909_110 |
0.2533157277 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 19 |
Hb_029223_040 |
0.2546914506 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 20 |
Hb_000580_110 |
0.2547178928 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |