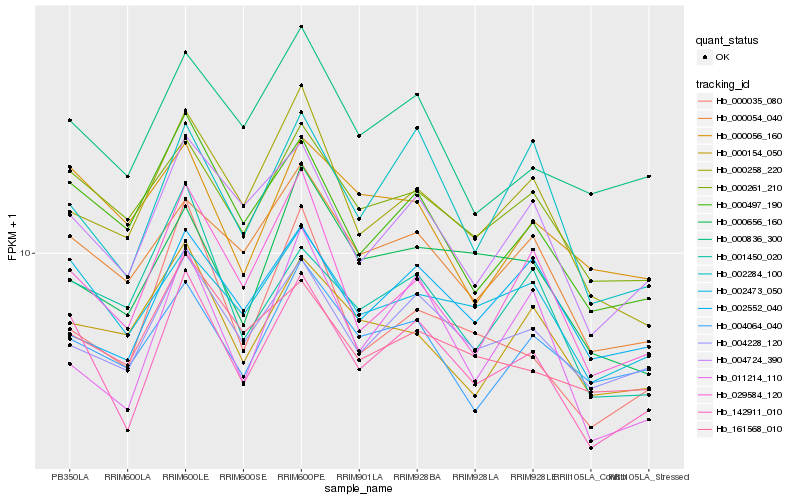
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_142911_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 2 |
Hb_004724_390 |
0.1061115669 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 3 |
Hb_000497_190 |
0.1112446184 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 4 |
Hb_002473_050 |
0.1157333744 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_004228_120 |
0.1177220841 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 6 |
Hb_000261_210 |
0.1189774699 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_029584_120 |
0.1193093994 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002284_100 |
0.119655738 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 9 |
Hb_000054_040 |
0.1203580648 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 10 |
Hb_000258_220 |
0.1280021558 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000836_300 |
0.1281761132 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 12 |
Hb_011214_110 |
0.1345816728 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 13 |
Hb_001450_020 |
0.1353150167 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000035_080 |
0.135577883 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 15 |
Hb_161568_010 |
0.1363013527 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 16 |
Hb_004064_040 |
0.1377835848 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 17 |
Hb_000656_160 |
0.138551038 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 18 |
Hb_000154_050 |
0.1424300334 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 19 |
Hb_002552_040 |
0.142956432 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 20 |
Hb_000056_160 |
0.14401972 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |