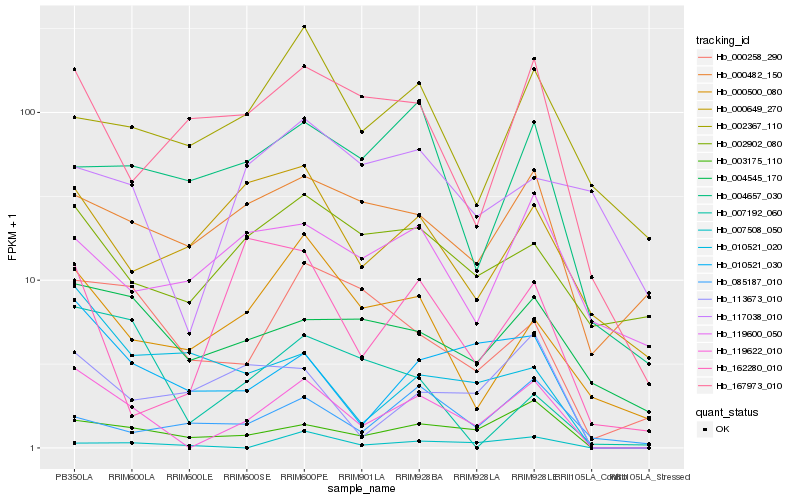
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119622_010 |
0.0 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 2 |
Hb_003175_110 |
0.2326598842 |
- |
- |
- |
| 3 |
Hb_010521_030 |
0.2455763401 |
- |
- |
- |
| 4 |
Hb_004545_170 |
0.2865120347 |
- |
- |
PREDICTED: autophagy-related protein 18h [Jatropha curcas] |
| 5 |
Hb_113673_010 |
0.2877656484 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 6 |
Hb_000258_290 |
0.2877870247 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 7 |
Hb_167973_010 |
0.2882461381 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 8 |
Hb_000500_080 |
0.2896107499 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 9 |
Hb_000482_150 |
0.2926424171 |
- |
- |
PREDICTED: zinc transporter 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002902_080 |
0.2943458247 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 11 |
Hb_000649_270 |
0.3050272083 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_004657_030 |
0.3073222444 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 13 |
Hb_002367_110 |
0.3090096198 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 14 |
Hb_007508_050 |
0.3111149559 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_119600_050 |
0.3123267759 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_010521_020 |
0.3147062128 |
- |
- |
- |
| 17 |
Hb_085187_010 |
0.3156669228 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_117038_010 |
0.3173387785 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 19 |
Hb_162280_010 |
0.3183597848 |
- |
- |
Phosphoprotein phosphatase isoform 2, partial [Theobroma cacao] |
| 20 |
Hb_007192_060 |
0.3191919904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |