| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
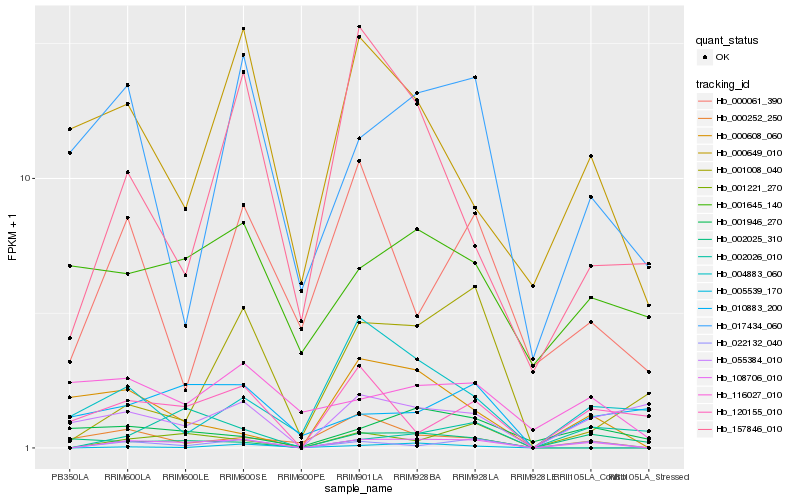
Hb_108706_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 2 |
Hb_022132_040 |
0.204114605 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 3 |
Hb_001221_270 |
0.3207010421 |
- |
- |
- |
| 4 |
Hb_120155_010 |
0.3345101247 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_017434_060 |
0.3389911969 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 6 |
Hb_002025_310 |
0.3393068874 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_055384_010 |
0.3413641962 |
- |
- |
- |
| 8 |
Hb_000252_250 |
0.3421305112 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_001946_270 |
0.3445336741 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 10 |
Hb_001008_040 |
0.3452475855 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 11 |
Hb_000649_010 |
0.3452642978 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 12 |
Hb_157846_010 |
0.3460308937 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 13 |
Hb_005539_170 |
0.3468324738 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 14 |
Hb_002026_010 |
0.3512909683 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 15 |
Hb_000608_060 |
0.3524147219 |
- |
- |
- |
| 16 |
Hb_001645_140 |
0.3538417424 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 17 |
Hb_010883_200 |
0.3540813686 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 18 |
Hb_004883_060 |
0.3561039525 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000061_390 |
0.3566506218 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 20 |
Hb_116027_010 |
0.3569361353 |
- |
- |
PREDICTED: separase [Jatropha curcas] |