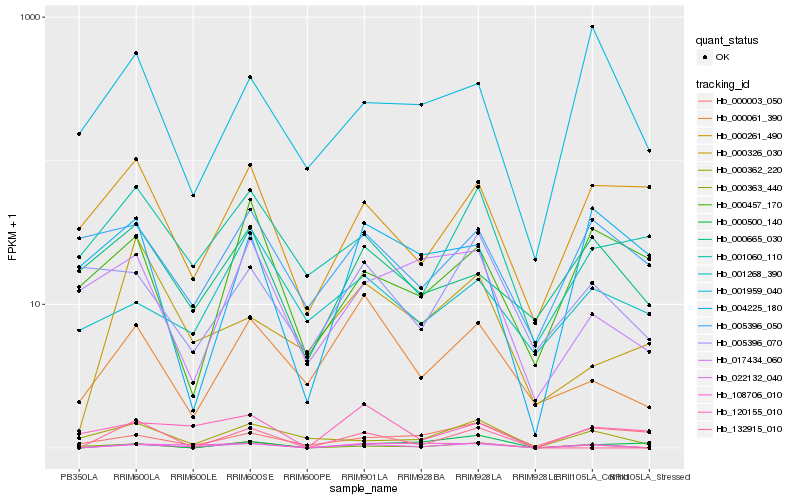
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022132_040 |
0.0 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 2 |
Hb_108706_010 |
0.204114605 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 3 |
Hb_000363_440 |
0.2658935616 |
- |
- |
- |
| 4 |
Hb_120155_010 |
0.3189599756 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_000061_390 |
0.3225838048 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 6 |
Hb_000362_220 |
0.3258765816 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 7 |
Hb_001959_040 |
0.3360726723 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000665_030 |
0.3466235599 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 9 |
Hb_017434_060 |
0.3519097427 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 10 |
Hb_001060_110 |
0.3533020624 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 11 |
Hb_005396_070 |
0.3546307904 |
- |
- |
hypothetical protein EUGRSUZ_B03067 [Eucalyptus grandis] |
| 12 |
Hb_000003_050 |
0.356461298 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 13 |
Hb_000261_490 |
0.3569645533 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 14 |
Hb_005396_050 |
0.3577875961 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 15 |
Hb_004225_180 |
0.3598071425 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 16 |
Hb_001268_390 |
0.3600934505 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 17 |
Hb_000500_140 |
0.3624190184 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 18 |
Hb_000457_170 |
0.3625338517 |
- |
- |
PREDICTED: uncharacterized protein LOC105633066 [Jatropha curcas] |
| 19 |
Hb_132915_010 |
0.3640134886 |
- |
- |
PREDICTED: uncharacterized protein LOC103700431, partial [Phoenix dactylifera] |
| 20 |
Hb_000326_030 |
0.3653502343 |
- |
- |
hypothetical protein PRUPE_ppa013909mg [Prunus persica] |