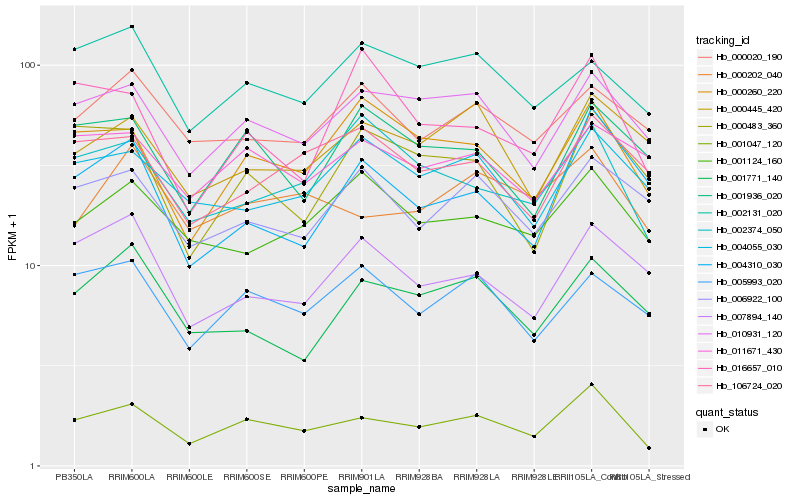
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001047_120 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 2 |
Hb_010931_120 |
0.1097238121 |
- |
- |
myosin heavy chain-related family protein [Populus trichocarpa] |
| 3 |
Hb_000260_220 |
0.1180767344 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000202_040 |
0.1289610232 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 5 |
Hb_005993_020 |
0.1304737766 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 6 |
Hb_002374_050 |
0.1308510872 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 7 |
Hb_004055_030 |
0.1367057448 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 8 |
Hb_001936_020 |
0.1369052227 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 9 |
Hb_001124_160 |
0.1370638092 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 10 |
Hb_006922_100 |
0.137951557 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X1 [Jatropha curcas] |
| 11 |
Hb_106724_020 |
0.138159887 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 1A-like isoform X1 [Populus euphratica] |
| 12 |
Hb_000445_420 |
0.1389236146 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_000483_360 |
0.1390793076 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 14 |
Hb_011671_430 |
0.1392909783 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 15 |
Hb_004310_030 |
0.1405387625 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 16 |
Hb_002131_020 |
0.1410869902 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 17 |
Hb_001771_140 |
0.1419255139 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 18 |
Hb_007894_140 |
0.1427836427 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 19 |
Hb_000020_190 |
0.1441369457 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 20 |
Hb_016657_010 |
0.1451877596 |
- |
- |
PREDICTED: peroxisome biogenesis protein 5 [Jatropha curcas] |