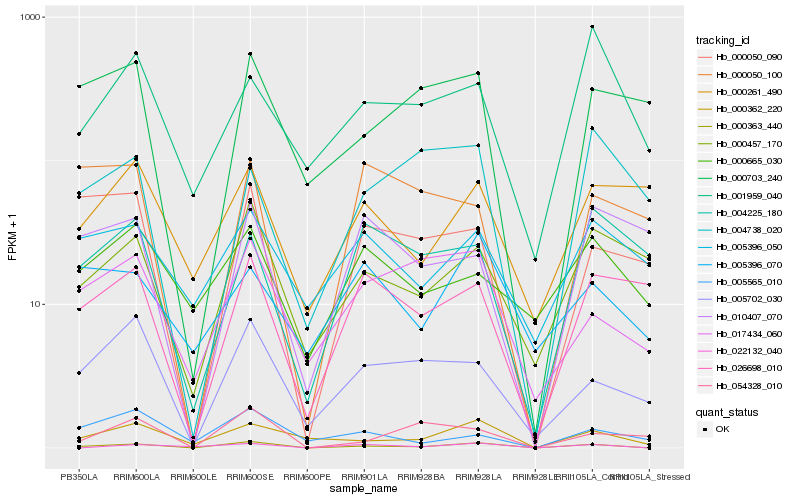
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_440 |
0.0 |
- |
- |
- |
| 2 |
Hb_000362_220 |
0.2563844713 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 3 |
Hb_022132_040 |
0.2658935616 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 4 |
Hb_005702_030 |
0.2721366048 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 5 |
Hb_000457_170 |
0.2789159375 |
- |
- |
PREDICTED: uncharacterized protein LOC105633066 [Jatropha curcas] |
| 6 |
Hb_017434_060 |
0.2813176674 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 7 |
Hb_000050_090 |
0.286108849 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 8 |
Hb_026698_010 |
0.2938519533 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_000703_240 |
0.2940388676 |
- |
- |
- |
| 10 |
Hb_054328_010 |
0.2959393029 |
- |
- |
hypothetical protein POPTR_0019s09820g [Populus trichocarpa] |
| 11 |
Hb_004738_020 |
0.3011488643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001959_040 |
0.3111216008 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005565_010 |
0.3137834184 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 14 |
Hb_004225_180 |
0.3138515319 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 15 |
Hb_005396_070 |
0.31801467 |
- |
- |
hypothetical protein EUGRSUZ_B03067 [Eucalyptus grandis] |
| 16 |
Hb_005396_050 |
0.320021763 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 17 |
Hb_000050_100 |
0.3204758096 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 2 [Jatropha curcas] |
| 18 |
Hb_010407_070 |
0.3237494089 |
- |
- |
hypothetical protein POPTR_0008s16750g [Populus trichocarpa] |
| 19 |
Hb_000261_490 |
0.3269318412 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 20 |
Hb_000665_030 |
0.3283257323 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |