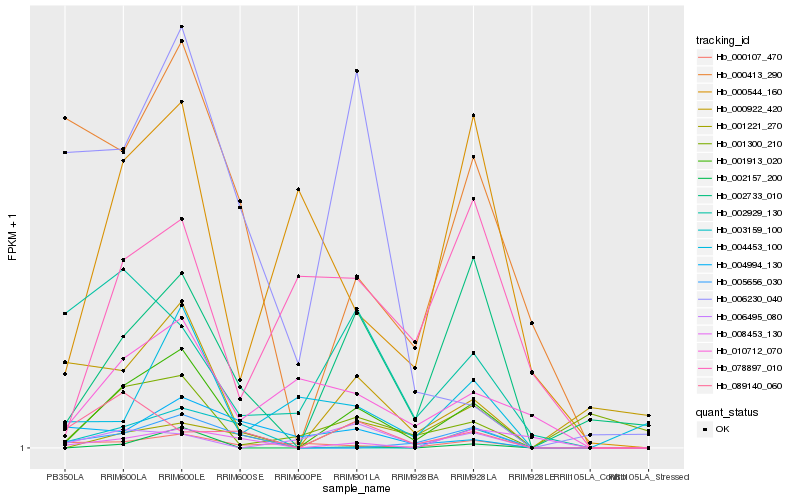
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001913_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002733_010 |
0.2464797854 |
- |
- |
- |
| 3 |
Hb_001221_270 |
0.3101679499 |
- |
- |
- |
| 4 |
Hb_005656_030 |
0.3273091819 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000413_290 |
0.3298689281 |
- |
- |
- |
| 6 |
Hb_010712_070 |
0.3344573123 |
- |
- |
Galactoside 2-alpha-L-fucosyltransferase [Aegilops tauschii] |
| 7 |
Hb_002157_200 |
0.3370532308 |
- |
- |
hypothetical protein VITISV_040170 [Vitis vinifera] |
| 8 |
Hb_000107_470 |
0.3395712655 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 9 |
Hb_000544_160 |
0.3539572903 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |
| 10 |
Hb_089140_060 |
0.3553265131 |
- |
- |
Uncharacterized protein TCM_037529 [Theobroma cacao] |
| 11 |
Hb_003159_100 |
0.3562306025 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002929_130 |
0.3576179783 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 13 |
Hb_000922_420 |
0.3608712307 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 14 |
Hb_004994_130 |
0.3654270595 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 15 |
Hb_078897_010 |
0.3657689027 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 16 |
Hb_008453_130 |
0.3665305807 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 17 |
Hb_001300_210 |
0.3688744036 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 18 |
Hb_006495_080 |
0.3701451793 |
- |
- |
hypothetical protein PRUPE_ppb025438mg, partial [Prunus persica] |
| 19 |
Hb_004453_100 |
0.3709043103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006230_040 |
0.3720166139 |
- |
- |
kinase, putative [Ricinus communis] |