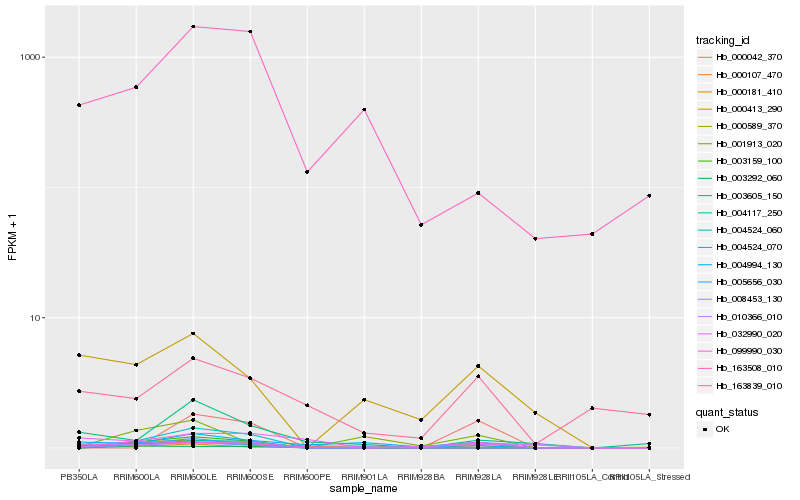
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003159_100 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_005656_030 |
0.1874478353 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 3 |
Hb_004524_070 |
0.3144779839 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 4 |
Hb_000413_290 |
0.3353528411 |
- |
- |
- |
| 5 |
Hb_000589_370 |
0.3557531446 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 6 |
Hb_001913_020 |
0.3562306025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_032990_020 |
0.3583290694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004524_060 |
0.3585673398 |
- |
- |
hypothetical protein VITISV_039334 [Vitis vinifera] |
| 9 |
Hb_004117_250 |
0.3606157888 |
- |
- |
PREDICTED: expansin-A8-like [Populus euphratica] |
| 10 |
Hb_000181_410 |
0.3618783941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008453_130 |
0.3621137734 |
- |
- |
hypothetical protein JCGZ_08179 [Jatropha curcas] |
| 12 |
Hb_003605_150 |
0.3629344184 |
- |
- |
chitinase family protein [Populus trichocarpa] |
| 13 |
Hb_000107_470 |
0.3660263542 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 14 |
Hb_000042_370 |
0.3691122737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_163508_010 |
0.3766044703 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 16 |
Hb_099990_030 |
0.3777477069 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_010366_010 |
0.3783819873 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 18 |
Hb_003292_060 |
0.3811996805 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38) [Jatropha curcas] |
| 19 |
Hb_163839_010 |
0.3829151689 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_004994_130 |
0.3929689326 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |