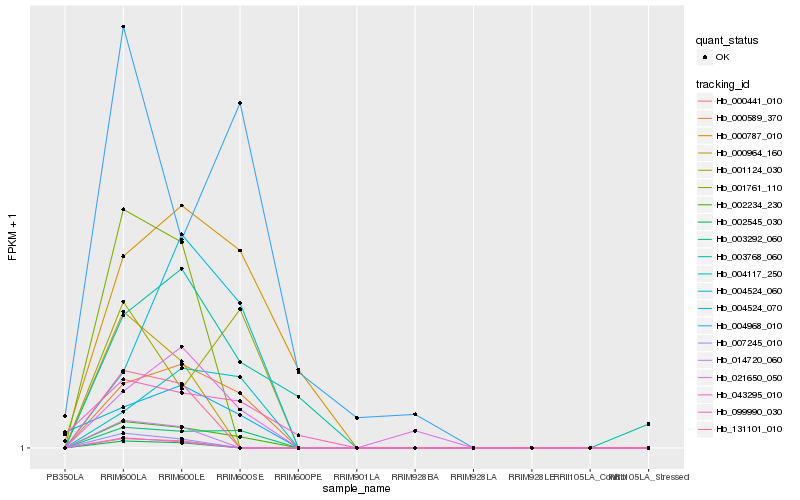
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002234_230 |
0.0 |
- |
- |
hypothetical protein JCGZ_26141 [Jatropha curcas] |
| 2 |
Hb_003292_060 |
0.1254071465 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38) [Jatropha curcas] |
| 3 |
Hb_000589_370 |
0.1265031913 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 4 |
Hb_001124_030 |
0.2344251134 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 5 |
Hb_021650_050 |
0.2565172154 |
- |
- |
hypothetical protein RCOM_0852210 [Ricinus communis] |
| 6 |
Hb_004117_250 |
0.2603351588 |
- |
- |
PREDICTED: expansin-A8-like [Populus euphratica] |
| 7 |
Hb_004524_070 |
0.2658491156 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 8 |
Hb_099990_030 |
0.2715433357 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_004524_060 |
0.2771867602 |
- |
- |
hypothetical protein VITISV_039334 [Vitis vinifera] |
| 10 |
Hb_000787_010 |
0.2785826512 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 11 |
Hb_003768_060 |
0.3000373627 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 12 |
Hb_004968_010 |
0.3095783714 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X1 [Populus euphratica] |
| 13 |
Hb_014720_060 |
0.3202092651 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 14 |
Hb_000441_010 |
0.3202140785 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 15 |
Hb_002545_030 |
0.3202857898 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_131101_010 |
0.3204505605 |
- |
- |
- |
| 17 |
Hb_001761_110 |
0.3206398072 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 18 |
Hb_000964_160 |
0.3235540892 |
- |
- |
PREDICTED: uncharacterized protein LOC102617979 isoform X2 [Citrus sinensis] |
| 19 |
Hb_007245_010 |
0.324036194 |
- |
- |
PREDICTED: uncharacterized protein LOC104783698 [Camelina sativa] |
| 20 |
Hb_043295_010 |
0.3240508666 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |