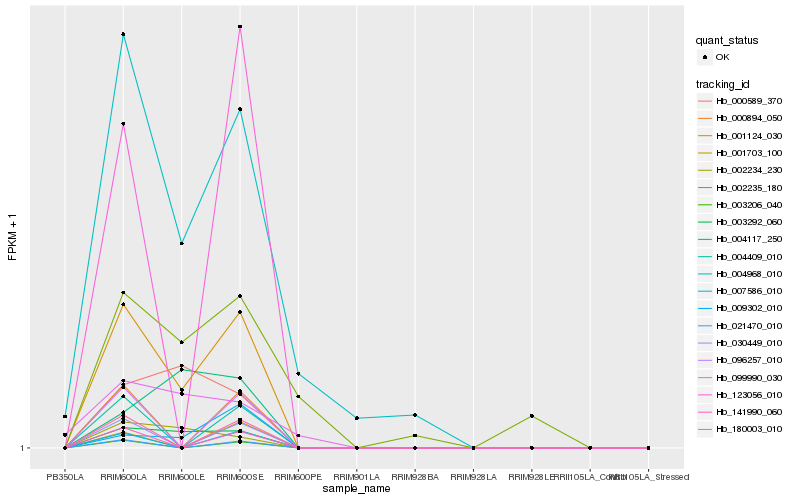
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 2 |
Hb_003292_060 |
0.1442040319 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38) [Jatropha curcas] |
| 3 |
Hb_002234_230 |
0.2344251134 |
- |
- |
hypothetical protein JCGZ_26141 [Jatropha curcas] |
| 4 |
Hb_009302_010 |
0.2399179949 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_000589_370 |
0.2433850626 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 6 |
Hb_004968_010 |
0.254049674 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X1 [Populus euphratica] |
| 7 |
Hb_004117_250 |
0.2845623799 |
- |
- |
PREDICTED: expansin-A8-like [Populus euphratica] |
| 8 |
Hb_000894_050 |
0.2939532969 |
- |
- |
- |
| 9 |
Hb_096257_010 |
0.2939770106 |
- |
- |
- |
| 10 |
Hb_180003_010 |
0.2944109941 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 11 |
Hb_030449_010 |
0.2944788849 |
- |
- |
- |
| 12 |
Hb_021470_010 |
0.2949500536 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_004409_010 |
0.295316961 |
- |
- |
- |
| 14 |
Hb_141990_060 |
0.2955537142 |
- |
- |
- |
| 15 |
Hb_003206_040 |
0.2956475787 |
- |
- |
Auxin efflux carrier component, putative [Ricinus communis] |
| 16 |
Hb_007586_010 |
0.2966880527 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 17 |
Hb_123056_010 |
0.3053471604 |
- |
- |
- |
| 18 |
Hb_099990_030 |
0.3082908064 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_001703_100 |
0.315605439 |
- |
- |
PREDICTED: uncharacterized protein LOC104885230 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_002235_180 |
0.3179744842 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |