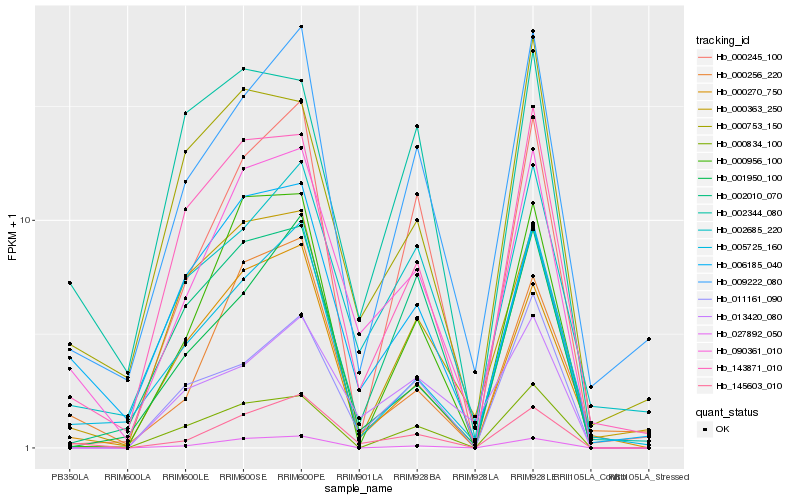
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090361_010 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_145603_010 |
0.0928542672 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 3 |
Hb_143871_010 |
0.1471000999 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_006185_040 |
0.1490827451 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 5 |
Hb_009222_080 |
0.1557499638 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 6 |
Hb_002685_220 |
0.15694319 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005725_160 |
0.1649587877 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 8 |
Hb_000956_100 |
0.1657738851 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 9 |
Hb_000270_750 |
0.168933325 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 10 |
Hb_000753_150 |
0.1690824954 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 11 |
Hb_000834_100 |
0.169971567 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 12 |
Hb_000256_220 |
0.1706134287 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 13 |
Hb_002010_070 |
0.1731007076 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 14 |
Hb_027892_050 |
0.17574596 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 15 |
Hb_002344_080 |
0.1786233431 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 16 |
Hb_011161_090 |
0.1792532344 |
- |
- |
- |
| 17 |
Hb_000245_100 |
0.1793021384 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 18 |
Hb_001950_100 |
0.181205511 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 19 |
Hb_013420_080 |
0.1822976574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000363_250 |
0.1825628578 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |