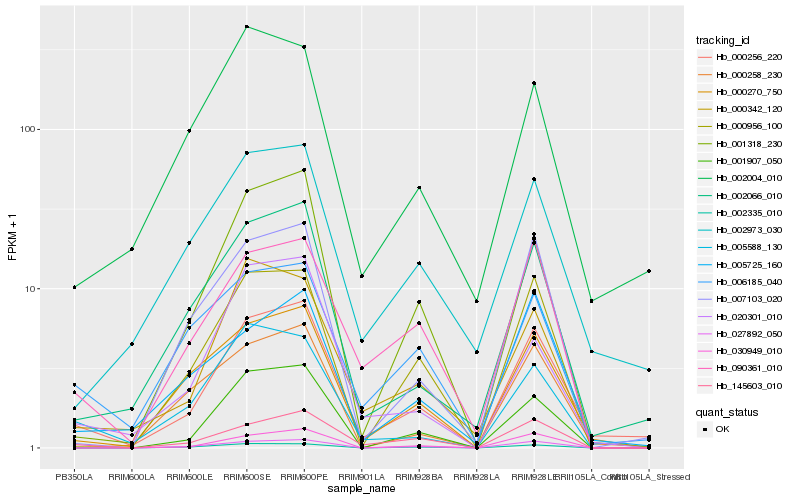
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_220 |
0.0 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 2 |
Hb_002066_010 |
0.1378265553 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000270_750 |
0.1453771268 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 4 |
Hb_145603_010 |
0.1472089769 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_002973_030 |
0.1606213364 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 6 |
Hb_001318_230 |
0.1616991185 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 7 |
Hb_007103_020 |
0.1653325733 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 8 |
Hb_030949_010 |
0.167864529 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 9 |
Hb_027892_050 |
0.1694965104 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 10 |
Hb_000956_100 |
0.1699818767 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 11 |
Hb_090361_010 |
0.1706134287 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_001907_050 |
0.1712630998 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 13 |
Hb_005725_160 |
0.171603326 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 14 |
Hb_000342_120 |
0.1768396252 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 15 |
Hb_006185_040 |
0.1778728217 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 16 |
Hb_002004_010 |
0.1795376162 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 17 |
Hb_000258_230 |
0.1799942252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005588_130 |
0.1809795617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002335_010 |
0.1837281027 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 20 |
Hb_020301_010 |
0.184144094 |
- |
- |
cytochrome P450, putative [Ricinus communis] |