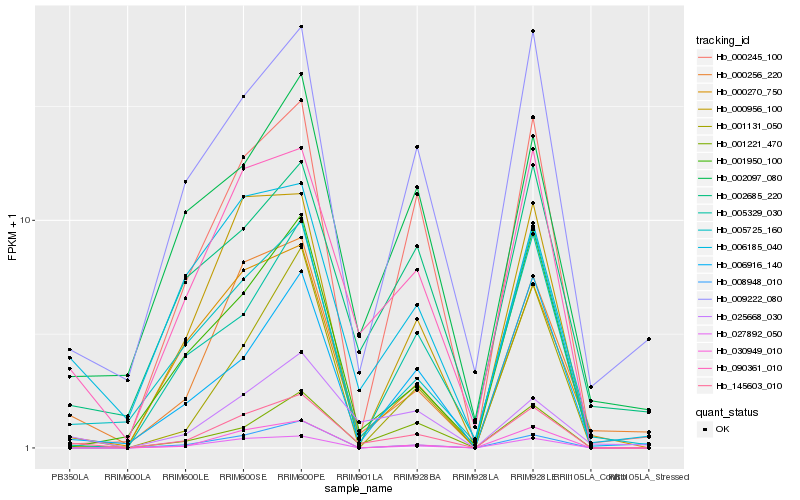
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145603_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_090361_010 |
0.0928542672 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_000256_220 |
0.1472089769 |
- |
- |
PREDICTED: flotillin-like protein 4 [Jatropha curcas] |
| 4 |
Hb_009222_080 |
0.1643803242 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 5 |
Hb_000245_100 |
0.1651037952 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 6 |
Hb_006185_040 |
0.1683862797 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 7 |
Hb_000270_750 |
0.1698781375 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 8 |
Hb_006916_140 |
0.1703207065 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 9 |
Hb_005725_160 |
0.1721035595 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 10 |
Hb_027892_050 |
0.1724837752 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 11 |
Hb_000956_100 |
0.174588374 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 12 |
Hb_001950_100 |
0.174652423 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 13 |
Hb_001221_470 |
0.1750782481 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 14 |
Hb_002097_080 |
0.1759470446 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 15 |
Hb_001131_050 |
0.1771985571 |
- |
- |
hypothetical protein EUGRSUZ_C02968 [Eucalyptus grandis] |
| 16 |
Hb_025668_030 |
0.1780075702 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 17 |
Hb_030949_010 |
0.1807575139 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 18 |
Hb_002685_220 |
0.1822033605 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005329_030 |
0.1828083854 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 20 |
Hb_008948_010 |
0.184592616 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |