| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
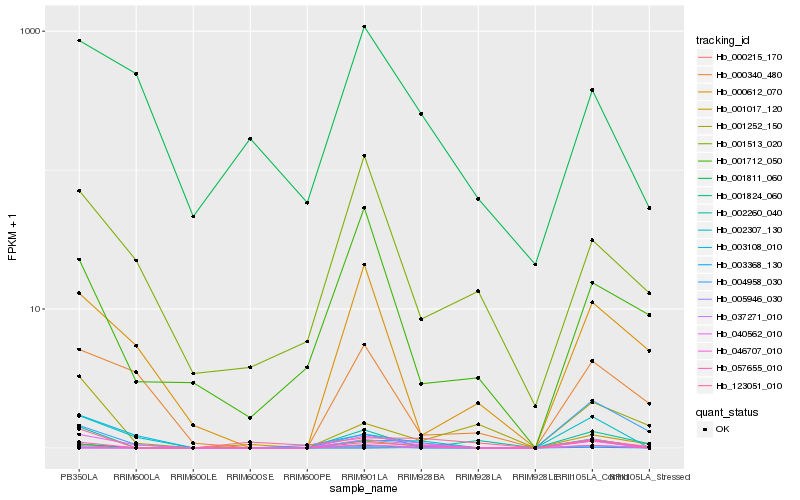
Hb_057655_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_040562_010 |
0.2515049753 |
- |
- |
- |
| 3 |
Hb_001824_060 |
0.2954199454 |
- |
- |
- |
| 4 |
Hb_002307_130 |
0.3216701284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002260_040 |
0.3779977312 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 6 |
Hb_000215_170 |
0.3799949312 |
- |
- |
PREDICTED: uncharacterized protein LOC102596491 [Solanum tuberosum] |
| 7 |
Hb_001017_120 |
0.3831584297 |
- |
- |
- |
| 8 |
Hb_037271_010 |
0.3975376867 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 9 |
Hb_004958_030 |
0.3993181972 |
- |
- |
PREDICTED: uncharacterized protein LOC104904188 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_123051_010 |
0.4042281023 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Populus euphratica] |
| 11 |
Hb_000612_070 |
0.4084067253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_046707_010 |
0.409918731 |
- |
- |
PREDICTED: uncharacterized protein LOC104887079 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_000340_480 |
0.4112318321 |
- |
- |
- |
| 14 |
Hb_003108_010 |
0.4130042403 |
- |
- |
- |
| 15 |
Hb_001252_150 |
0.4143847375 |
- |
- |
- |
| 16 |
Hb_003368_130 |
0.4165518411 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_001712_050 |
0.4181091563 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 18 |
Hb_001811_060 |
0.4271658685 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005946_030 |
0.4280808348 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 20 |
Hb_001513_020 |
0.4311040378 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |