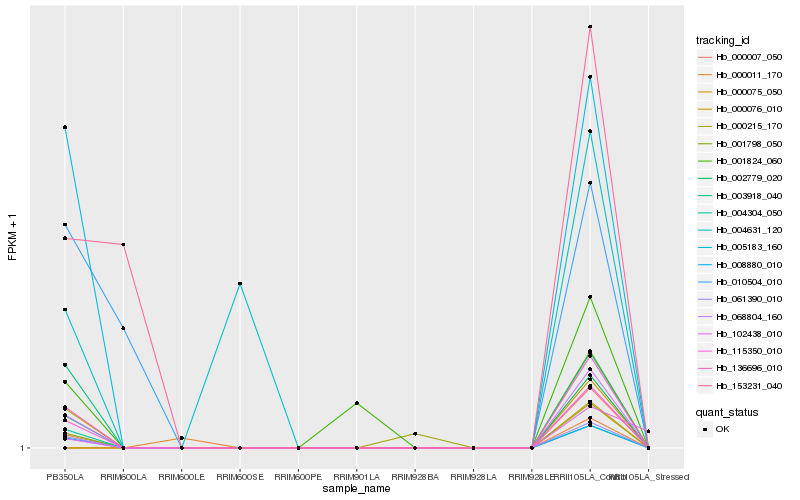
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_160 |
0.0 |
transcription factor |
TF Family: MIKC |
hypothetical protein JCGZ_12632 [Jatropha curcas] |
| 2 |
Hb_136696_010 |
0.0127151777 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_002779_020 |
0.0135531687 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_115350_010 |
0.0140042705 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 5 |
Hb_061390_010 |
0.0155466925 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 6 |
Hb_008880_010 |
0.0156762976 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 isoform X2 [Nelumbo nucifera] |
| 7 |
Hb_004304_050 |
0.1435487255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 8 |
Hb_005183_160 |
0.1480087758 |
- |
- |
- |
| 9 |
Hb_003918_040 |
0.1543405779 |
- |
- |
- |
| 10 |
Hb_001798_050 |
0.170305887 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 11 |
Hb_001824_060 |
0.2971565942 |
- |
- |
- |
| 12 |
Hb_000215_170 |
0.3141656004 |
- |
- |
PREDICTED: uncharacterized protein LOC102596491 [Solanum tuberosum] |
| 13 |
Hb_000011_170 |
0.3153846918 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 14 |
Hb_010504_010 |
0.3448569954 |
- |
- |
40S ribosomal protein S15C [Hevea brasiliensis] |
| 15 |
Hb_153231_040 |
0.3549409973 |
- |
- |
- |
| 16 |
Hb_004631_120 |
0.3758213115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_102438_010 |
0.379856341 |
- |
- |
- |
| 18 |
Hb_000007_050 |
0.4027186473 |
- |
- |
- |
| 19 |
Hb_000075_050 |
0.4027186473 |
- |
- |
- |
| 20 |
Hb_000076_010 |
0.4027186473 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |