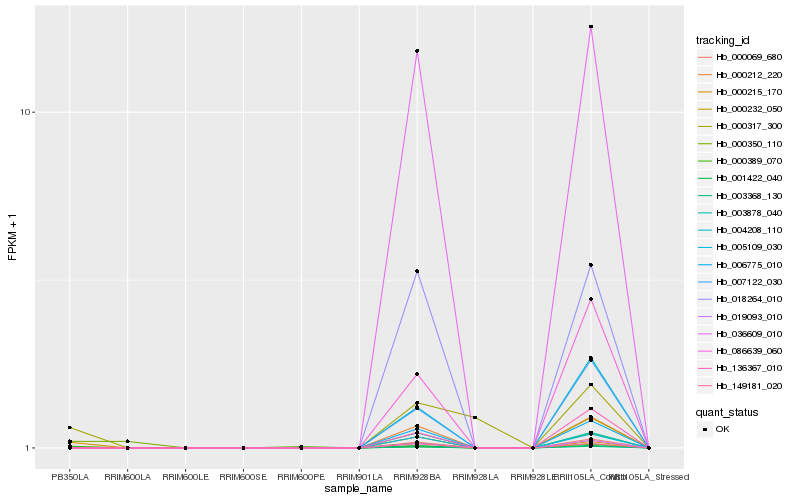
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003368_130 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_000215_170 |
0.2040069873 |
- |
- |
PREDICTED: uncharacterized protein LOC102596491 [Solanum tuberosum] |
| 3 |
Hb_000389_070 |
0.308569076 |
- |
- |
- |
| 4 |
Hb_000317_300 |
0.3195200288 |
- |
- |
- |
| 5 |
Hb_019093_010 |
0.3408737406 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 6 |
Hb_000069_680 |
0.3408920601 |
- |
- |
- |
| 7 |
Hb_006775_010 |
0.3412286338 |
transcription factor |
TF Family: M-type |
AGAMOUS [Populus yunnanensis] |
| 8 |
Hb_000212_220 |
0.3414007355 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 9 |
Hb_000350_110 |
0.343972763 |
- |
- |
PREDICTED: uncharacterized protein LOC104220461 isoform X2 [Nicotiana sylvestris] |
| 10 |
Hb_005109_030 |
0.3440289096 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 11 |
Hb_036609_010 |
0.3457531312 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 12 |
Hb_001422_040 |
0.3501852376 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 13 |
Hb_018264_010 |
0.3503037097 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 14 |
Hb_149181_020 |
0.3504193425 |
- |
- |
- |
| 15 |
Hb_003878_040 |
0.3518640619 |
- |
- |
- |
| 16 |
Hb_004208_110 |
0.3537087584 |
- |
- |
- |
| 17 |
Hb_007122_030 |
0.3537382121 |
- |
- |
- |
| 18 |
Hb_086639_060 |
0.354290273 |
- |
- |
hypothetical protein JCGZ_25622 [Jatropha curcas] |
| 19 |
Hb_136367_010 |
0.3560131655 |
- |
- |
hypothetical protein POPTR_0012s02040g [Populus trichocarpa] |
| 20 |
Hb_000232_050 |
0.3584060947 |
- |
- |
- |