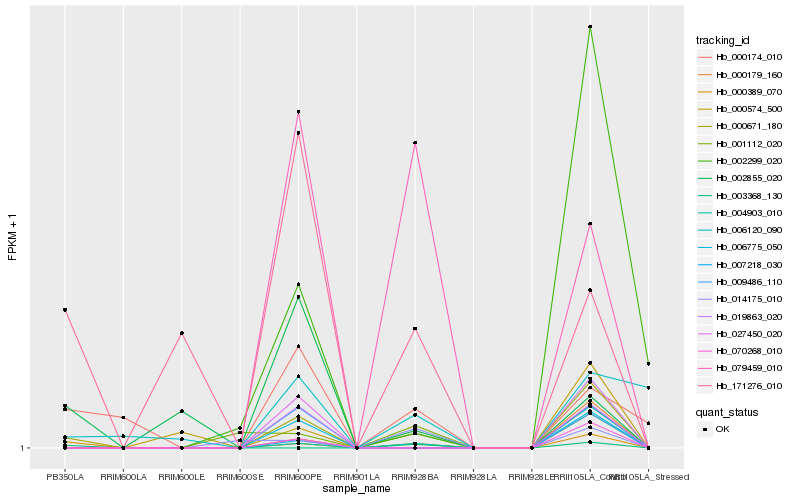
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_180 |
0.0 |
- |
- |
RING zinc finger protein, putative [Arabidopsis thaliana] |
| 2 |
Hb_000389_070 |
0.1858273476 |
- |
- |
- |
| 3 |
Hb_004903_010 |
0.3035592493 |
- |
- |
- |
| 4 |
Hb_019863_020 |
0.3283131678 |
- |
- |
PREDICTED: wound-induced protein 1 [Jatropha curcas] |
| 5 |
Hb_070268_010 |
0.3294832324 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 6 |
Hb_006775_050 |
0.3384991895 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29670 isoform X2 [Nicotiana sylvestris] |
| 7 |
Hb_000179_160 |
0.3588889298 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 8 |
Hb_007218_030 |
0.365272239 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Jatropha curcas] |
| 9 |
Hb_002299_020 |
0.3690877751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009486_110 |
0.3734947607 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 11 |
Hb_027450_020 |
0.3743142701 |
- |
- |
PREDICTED: B3 domain-containing protein At2g31420-like [Citrus sinensis] |
| 12 |
Hb_079459_010 |
0.3794754726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_171276_010 |
0.3962466696 |
- |
- |
- |
| 14 |
Hb_000174_010 |
0.4003861685 |
- |
- |
PREDICTED: uncharacterized protein LOC103444904 [Malus domestica] |
| 15 |
Hb_001112_020 |
0.404527761 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 16 |
Hb_000574_500 |
0.4197300348 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 17 |
Hb_014175_010 |
0.4233724024 |
- |
- |
Uncharacterized protein TCM_033221 [Theobroma cacao] |
| 18 |
Hb_003368_130 |
0.4266605567 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_006120_090 |
0.4290638527 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 20 |
Hb_002855_020 |
0.431100455 |
- |
- |
- |