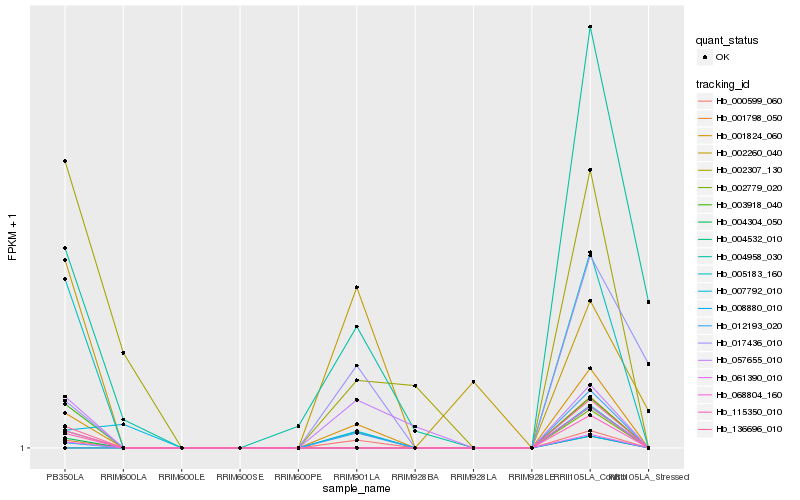
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001824_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_057655_010 |
0.2954199454 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_136696_010 |
0.2971409149 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 4 |
Hb_068804_160 |
0.2971565942 |
transcription factor |
TF Family: MIKC |
hypothetical protein JCGZ_12632 [Jatropha curcas] |
| 5 |
Hb_002779_020 |
0.2971572641 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_115350_010 |
0.2971669543 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 7 |
Hb_061390_010 |
0.2972047886 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 8 |
Hb_008880_010 |
0.2972082989 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_004958_030 |
0.3206722577 |
- |
- |
PREDICTED: uncharacterized protein LOC104904188 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_004304_050 |
0.3244264703 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 11 |
Hb_005183_160 |
0.3261573741 |
- |
- |
- |
| 12 |
Hb_003918_040 |
0.3286935986 |
- |
- |
- |
| 13 |
Hb_001798_050 |
0.3420550627 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 14 |
Hb_017436_010 |
0.3497117712 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_002307_130 |
0.3584493948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007792_010 |
0.3613243897 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 17 |
Hb_012193_020 |
0.3695749113 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_004532_010 |
0.3727029742 |
- |
- |
PREDICTED: uncharacterized protein LOC105800934 [Gossypium raimondii] |
| 19 |
Hb_000599_060 |
0.3771199293 |
- |
- |
PREDICTED: legumin A-like [Jatropha curcas] |
| 20 |
Hb_002260_040 |
0.3793854285 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |