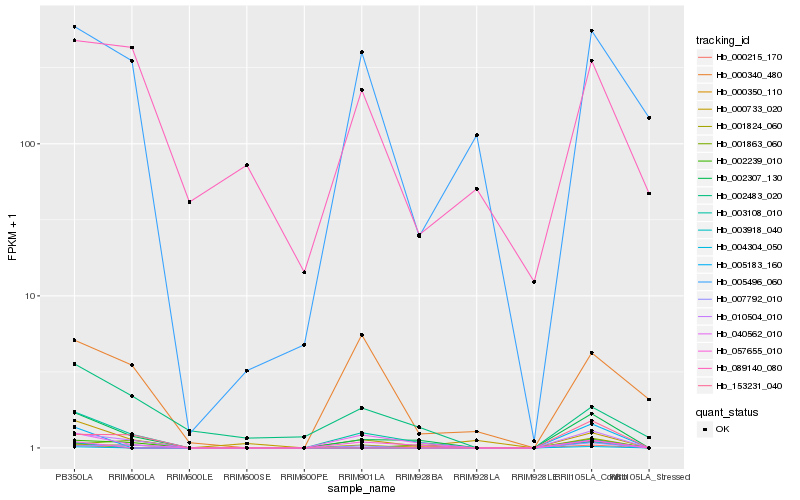
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010504_010 |
0.2846379551 |
- |
- |
40S ribosomal protein S15C [Hevea brasiliensis] |
| 3 |
Hb_000350_110 |
0.2883881154 |
- |
- |
PREDICTED: uncharacterized protein LOC104220461 isoform X2 [Nicotiana sylvestris] |
| 4 |
Hb_040562_010 |
0.2995015803 |
- |
- |
- |
| 5 |
Hb_003108_010 |
0.3013794591 |
- |
- |
- |
| 6 |
Hb_007792_010 |
0.3089600246 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 7 |
Hb_002239_010 |
0.3165681197 |
- |
- |
- |
| 8 |
Hb_000215_170 |
0.320919636 |
- |
- |
PREDICTED: uncharacterized protein LOC102596491 [Solanum tuberosum] |
| 9 |
Hb_057655_010 |
0.3216701284 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_000733_020 |
0.3218597038 |
- |
- |
- |
| 11 |
Hb_153231_040 |
0.3323384736 |
- |
- |
- |
| 12 |
Hb_005496_060 |
0.3379221957 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 38-like [Jatropha curcas] |
| 13 |
Hb_002483_020 |
0.3418592204 |
- |
- |
- |
| 14 |
Hb_001863_060 |
0.3444189582 |
- |
- |
PIF-like transposase [Daucus carota] |
| 15 |
Hb_001824_060 |
0.3584493948 |
- |
- |
- |
| 16 |
Hb_089140_080 |
0.3631223272 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003918_040 |
0.3694560191 |
- |
- |
- |
| 18 |
Hb_005183_160 |
0.3700788996 |
- |
- |
- |
| 19 |
Hb_000340_480 |
0.3704189532 |
- |
- |
- |
| 20 |
Hb_004304_050 |
0.3705734781 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |