| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
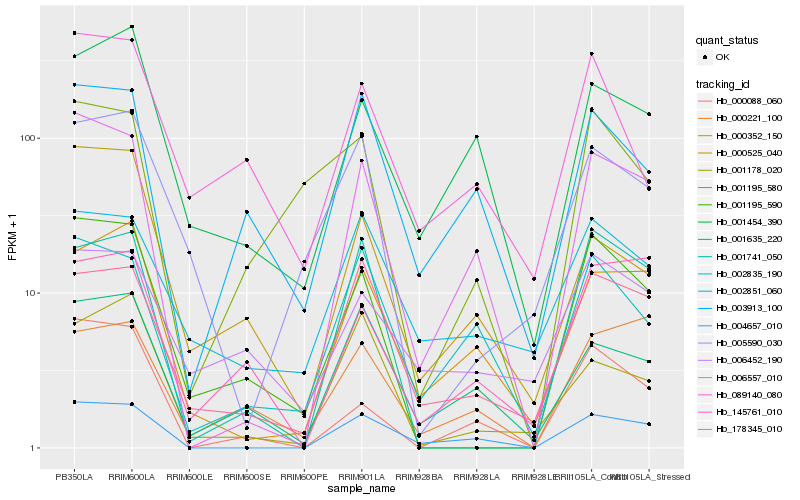
Hb_001195_590 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 2 |
Hb_001741_050 |
0.1669842822 |
- |
- |
- |
| 3 |
Hb_000088_060 |
0.1879204403 |
- |
- |
- |
| 4 |
Hb_005590_030 |
0.1892454062 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 5 |
Hb_001195_580 |
0.1967354139 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 6 |
Hb_006557_010 |
0.2027614031 |
- |
- |
- |
| 7 |
Hb_089140_080 |
0.2074745658 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 8 |
Hb_178345_010 |
0.212401288 |
- |
- |
hypothetical protein PHAVU_004G084200g [Phaseolus vulgaris] |
| 9 |
Hb_006452_190 |
0.2167397833 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_145761_010 |
0.2198042519 |
- |
- |
- |
| 11 |
Hb_000525_040 |
0.2213288649 |
- |
- |
- |
| 12 |
Hb_004657_010 |
0.2276157217 |
- |
- |
PREDICTED: uncharacterized protein LOC105639764 [Jatropha curcas] |
| 13 |
Hb_003913_100 |
0.2296695649 |
- |
- |
copper amine oxidase, putative [Ricinus communis] |
| 14 |
Hb_000352_150 |
0.2318600306 |
- |
- |
hypothetical protein JCGZ_11641 [Jatropha curcas] |
| 15 |
Hb_001454_390 |
0.2328029716 |
- |
- |
8-kd dynein light chain, DLC8, pin [Thalassiosira pseudonana CCMP1335] |
| 16 |
Hb_002851_060 |
0.2346175954 |
- |
- |
Clathrin light chain 2 -like protein [Gossypium arboreum] |
| 17 |
Hb_001178_020 |
0.2352478301 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 3 [Jatropha curcas] |
| 18 |
Hb_001635_220 |
0.2366022077 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 19 |
Hb_002835_190 |
0.2373360197 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 20 |
Hb_000221_100 |
0.2400240696 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |