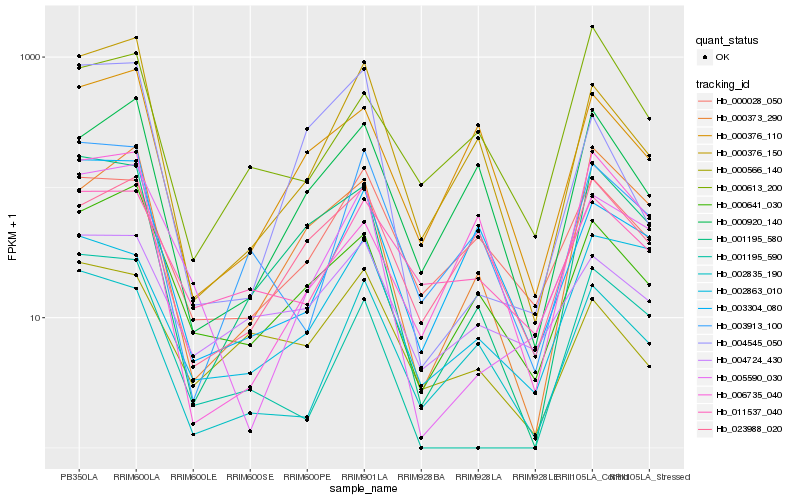
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_580 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s04130g [Populus trichocarpa] |
| 2 |
Hb_000373_290 |
0.1518747664 |
- |
- |
RecName: Full=Citrate-binding protein; Flags: Precursor [Hevea brasiliensis] |
| 3 |
Hb_000641_030 |
0.1791040398 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 4 |
Hb_002863_010 |
0.191785729 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 5 |
Hb_001195_590 |
0.1967354139 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 6 |
Hb_000376_110 |
0.1980953552 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Brachypodium distachyon] |
| 7 |
Hb_004724_430 |
0.200159235 |
- |
- |
PREDICTED: F-box protein At2g27310 [Jatropha curcas] |
| 8 |
Hb_023988_020 |
0.2052850794 |
- |
- |
PREDICTED: transmembrane protein 19 [Prunus mume] |
| 9 |
Hb_003913_100 |
0.2063990982 |
- |
- |
copper amine oxidase, putative [Ricinus communis] |
| 10 |
Hb_002835_190 |
0.2065440592 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 11 |
Hb_000376_150 |
0.207309942 |
- |
- |
O-acyltransferase WSD1 [Gossypium arboreum] |
| 12 |
Hb_000920_140 |
0.2091555817 |
- |
- |
annexin [Manihot esculenta] |
| 13 |
Hb_005590_030 |
0.2119877915 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_000028_050 |
0.2135926255 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 15 |
Hb_000566_140 |
0.2149642354 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 16 |
Hb_011537_040 |
0.2174421119 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 17 |
Hb_000613_200 |
0.2182329442 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 18 |
Hb_006735_040 |
0.2212281103 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_003304_080 |
0.2231781288 |
transcription factor |
TF Family: bHLH |
lMYC3 [Hevea brasiliensis] |
| 20 |
Hb_004545_050 |
0.2241740903 |
- |
- |
Cu/Zn superoxide dismutase [Hevea brasiliensis] |