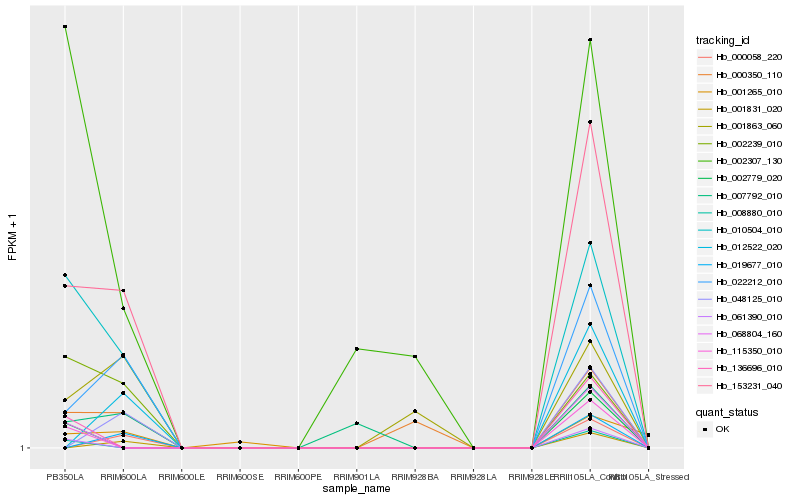
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153231_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_010504_010 |
0.1204869667 |
- |
- |
40S ribosomal protein S15C [Hevea brasiliensis] |
| 3 |
Hb_022212_010 |
0.1436860466 |
- |
- |
- |
| 4 |
Hb_002239_010 |
0.1889715063 |
- |
- |
- |
| 5 |
Hb_000350_110 |
0.2795621826 |
- |
- |
PREDICTED: uncharacterized protein LOC104220461 isoform X2 [Nicotiana sylvestris] |
| 6 |
Hb_001863_060 |
0.2873380527 |
- |
- |
PIF-like transposase [Daucus carota] |
| 7 |
Hb_007792_010 |
0.2998104276 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 8 |
Hb_002307_130 |
0.3323384736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008880_010 |
0.3545515669 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 isoform X2 [Nelumbo nucifera] |
| 10 |
Hb_061390_010 |
0.3545523134 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 11 |
Hb_115350_010 |
0.3545643635 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 12 |
Hb_002779_020 |
0.3545689918 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_136696_010 |
0.3545789158 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 14 |
Hb_068804_160 |
0.3549409973 |
transcription factor |
TF Family: MIKC |
hypothetical protein JCGZ_12632 [Jatropha curcas] |
| 15 |
Hb_001265_010 |
0.3560824721 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 16 |
Hb_048125_010 |
0.3609903668 |
- |
- |
PREDICTED: uncharacterized protein LOC104104928 [Nicotiana tomentosiformis] |
| 17 |
Hb_019677_010 |
0.3609986267 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000058_220 |
0.3610008642 |
- |
- |
hypothetical protein, partial [Staphylococcus aureus] |
| 19 |
Hb_012522_020 |
0.3610038901 |
- |
- |
- |
| 20 |
Hb_001831_020 |
0.3610090061 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |