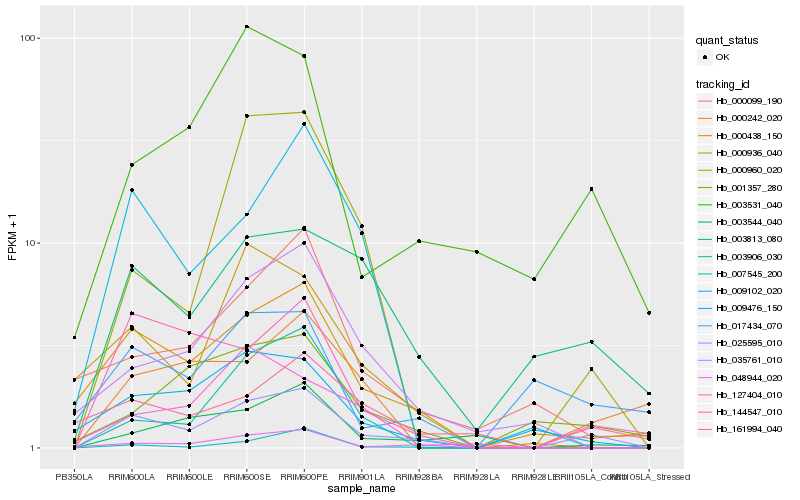
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025595_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 2 |
Hb_000438_150 |
0.2243099067 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 3 |
Hb_048944_020 |
0.2392269971 |
- |
- |
hypothetical protein POPTR_0014s002002g, partial [Populus trichocarpa] |
| 4 |
Hb_003813_080 |
0.2500761436 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 5 |
Hb_009476_150 |
0.2552597097 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 6 |
Hb_003531_040 |
0.2568853359 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 7 |
Hb_000960_020 |
0.2671959809 |
- |
- |
- |
| 8 |
Hb_001357_280 |
0.2683658611 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 9 |
Hb_003544_040 |
0.269744552 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 10 |
Hb_144547_010 |
0.2711074335 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_035761_010 |
0.2733195108 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 12 |
Hb_161994_040 |
0.274824014 |
- |
- |
- |
| 13 |
Hb_127404_010 |
0.2764117594 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X1 [Gossypium raimondii] |
| 14 |
Hb_009102_020 |
0.2773692775 |
- |
- |
hypothetical protein EUGRSUZ_I01262 [Eucalyptus grandis] |
| 15 |
Hb_003906_030 |
0.2816238391 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 16 |
Hb_000936_040 |
0.28319123 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 17 |
Hb_000242_020 |
0.2837637129 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 18 |
Hb_007545_200 |
0.2896593504 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 19 |
Hb_000099_190 |
0.2901668703 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 20 |
Hb_017434_070 |
0.2915240286 |
- |
- |
transcription factor, putative [Ricinus communis] |