| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
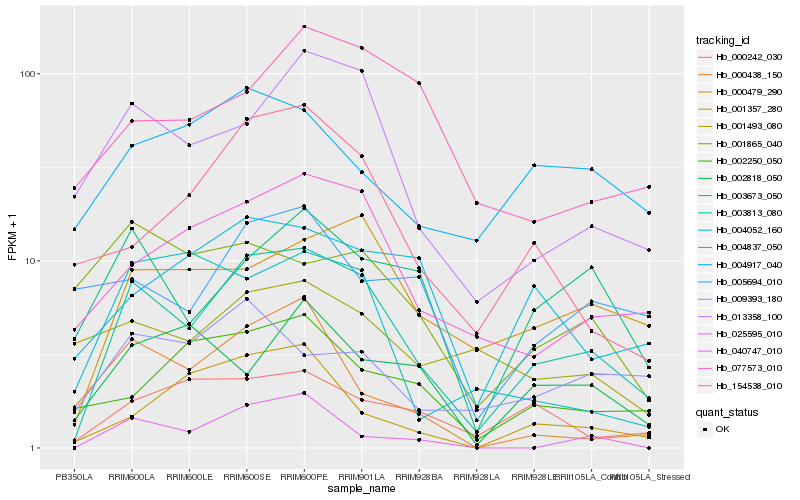
Hb_003813_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 2 |
Hb_040747_010 |
0.1960677932 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 3 |
Hb_013358_100 |
0.1981959444 |
- |
- |
unknown [Medicago truncatula] |
| 4 |
Hb_000479_290 |
0.2037248888 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003673_050 |
0.2170658336 |
- |
- |
hypothetical protein POPTR_0001s40990g [Populus trichocarpa] |
| 6 |
Hb_154538_010 |
0.2221741005 |
- |
- |
- |
| 7 |
Hb_000242_030 |
0.223677961 |
- |
- |
PREDICTED: uncharacterized protein LOC102608003 [Citrus sinensis] |
| 8 |
Hb_004837_050 |
0.2304659338 |
- |
- |
- |
| 9 |
Hb_002250_050 |
0.2406237106 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 10 |
Hb_009393_180 |
0.241409949 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_004917_040 |
0.2423354192 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 12 |
Hb_001357_280 |
0.2442955 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 13 |
Hb_025595_010 |
0.2500761436 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_077573_010 |
0.2509132315 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_005694_010 |
0.2515728817 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 16 |
Hb_002818_050 |
0.2545076218 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_004052_160 |
0.2548290751 |
- |
- |
Chain A, Rna Binding Protein |
| 18 |
Hb_001493_080 |
0.2558382693 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 19 |
Hb_000438_150 |
0.2559244307 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 20 |
Hb_001865_040 |
0.2569206858 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |