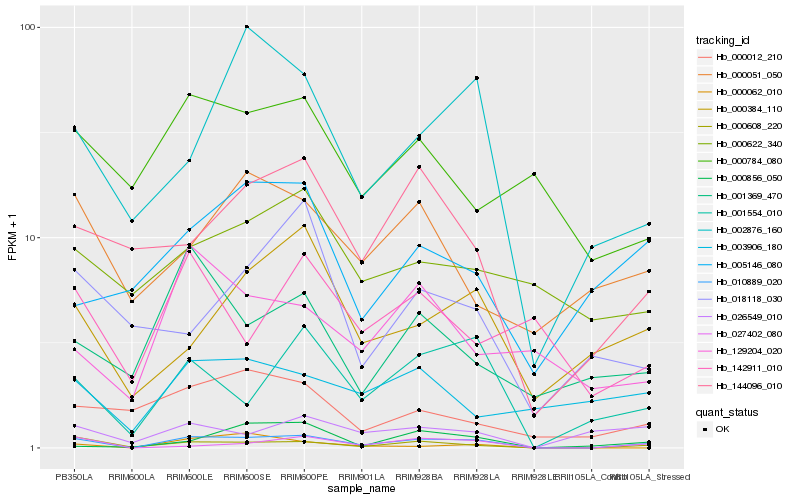
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010889_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000608_220 |
0.1873748178 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 3 |
Hb_027402_080 |
0.2349126565 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 4 |
Hb_001554_010 |
0.2366255844 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 5 |
Hb_144096_010 |
0.2697655628 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 6 |
Hb_002876_160 |
0.2742930556 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 7 |
Hb_000384_110 |
0.2818943324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_026549_010 |
0.2858571117 |
- |
- |
- |
| 9 |
Hb_000012_210 |
0.291961391 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000051_050 |
0.2989650679 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 11 |
Hb_001369_470 |
0.2993155467 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 12 |
Hb_129204_020 |
0.3060254486 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 13 |
Hb_003906_180 |
0.3064523563 |
- |
- |
unknown [Zea mays] |
| 14 |
Hb_000856_050 |
0.3071874954 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 15 |
Hb_005146_080 |
0.3094860888 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000784_080 |
0.3096896451 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 17 |
Hb_000062_010 |
0.3107265431 |
- |
- |
Alpha-L-fucosidase 2 precursor, putative [Ricinus communis] |
| 18 |
Hb_018118_030 |
0.3110815651 |
- |
- |
PREDICTED: putative multidrug resistance protein, partial [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_142911_010 |
0.3119041402 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g49730 [Jatropha curcas] |
| 20 |
Hb_000622_340 |
0.3121238772 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |