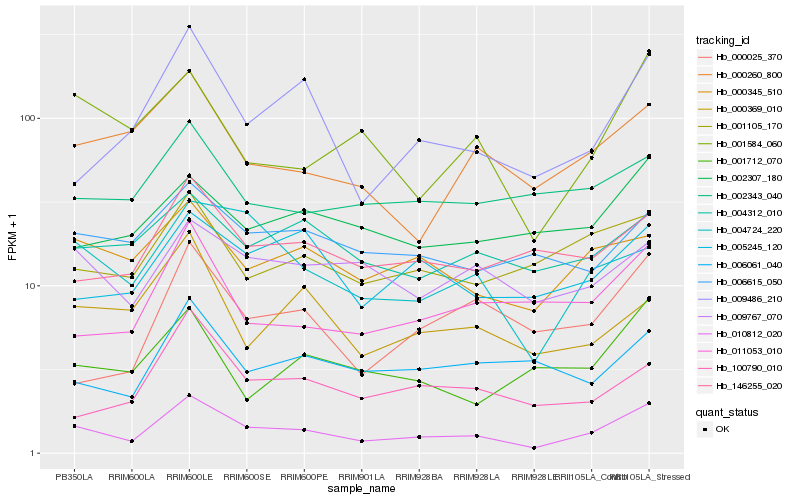
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_020 |
0.0 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 2 |
Hb_009486_210 |
0.1639207209 |
- |
- |
- |
| 3 |
Hb_004724_220 |
0.1674327861 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 4 |
Hb_001584_060 |
0.1715671897 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 5 |
Hb_000345_510 |
0.1739676339 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 6 |
Hb_000260_800 |
0.1761316685 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 7 |
Hb_011053_010 |
0.1762048148 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 8 |
Hb_001105_170 |
0.177031764 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005245_120 |
0.179138254 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_009767_070 |
0.1800596128 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 11 |
Hb_146255_020 |
0.1811293905 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 12 |
Hb_100790_010 |
0.1840028144 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004312_010 |
0.1849591608 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006615_050 |
0.1863878342 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 15 |
Hb_000025_370 |
0.1865955208 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006061_040 |
0.1871030871 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002307_180 |
0.1873110671 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002343_040 |
0.1886370467 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000369_010 |
0.189223959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001712_070 |
0.1893684855 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |