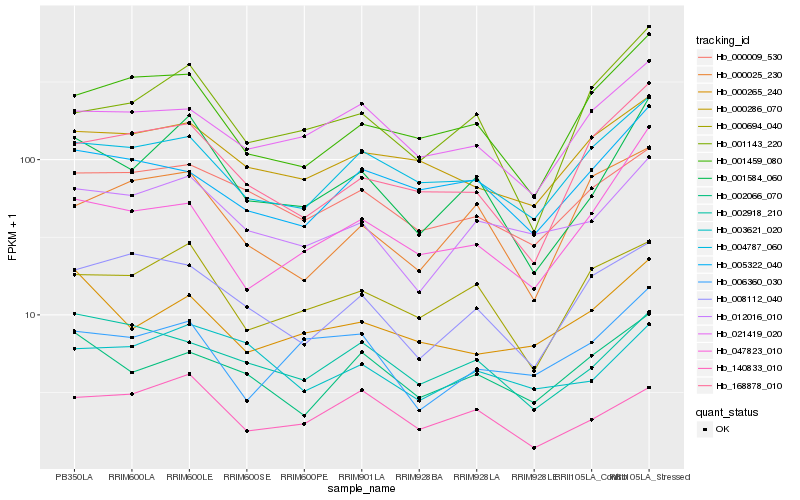
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_060 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 2 |
Hb_001143_220 |
0.1226757214 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 3 |
Hb_012016_010 |
0.1283187707 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000694_040 |
0.1311155705 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_001459_080 |
0.1345979726 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 6 |
Hb_168878_010 |
0.1378220568 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 7 |
Hb_004787_060 |
0.1378993356 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 8 |
Hb_140833_010 |
0.1430153146 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 9 |
Hb_002066_070 |
0.1491817039 |
- |
- |
PREDICTED: uncharacterized protein LOC105631043 [Jatropha curcas] |
| 10 |
Hb_047823_010 |
0.1500013898 |
- |
- |
hypothetical protein CARUB_v10024013mg [Capsella rubella] |
| 11 |
Hb_021419_020 |
0.1518605166 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 12 |
Hb_000009_530 |
0.1525336169 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 13 |
Hb_008112_040 |
0.1534727005 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 14 |
Hb_002918_210 |
0.1536511645 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 15 |
Hb_003621_020 |
0.1567338376 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 16 |
Hb_006360_030 |
0.1578451442 |
- |
- |
- |
| 17 |
Hb_005322_040 |
0.1578499408 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 18 |
Hb_000286_070 |
0.1579199473 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 19 |
Hb_000265_240 |
0.1587318109 |
- |
- |
PREDICTED: uncharacterized protein LOC105643590 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000025_230 |
0.1595354438 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |