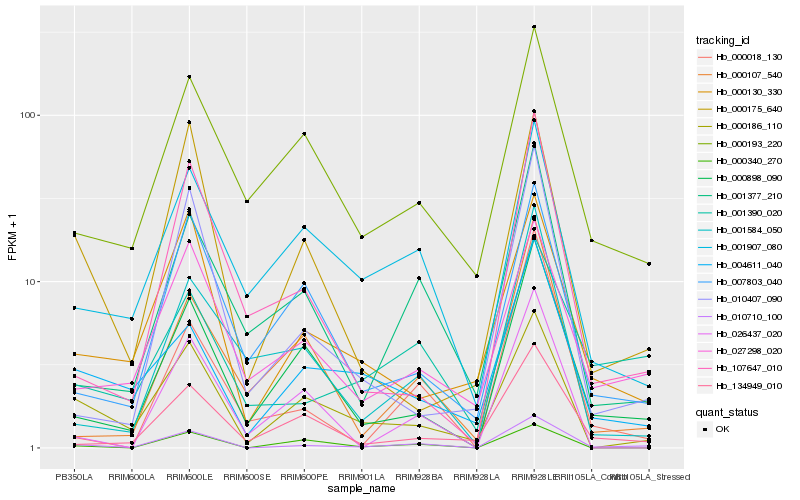
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010710_100 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_000898_090 |
0.186337223 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105633600 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000186_110 |
0.2032123556 |
- |
- |
hypothetical protein CISIN_1g020639mg [Citrus sinensis] |
| 4 |
Hb_001377_210 |
0.2045806534 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_027298_020 |
0.2110791012 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 6 |
Hb_107647_010 |
0.2169828601 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 7 |
Hb_010407_090 |
0.2182569175 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 8 |
Hb_001390_020 |
0.2195363776 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_000018_130 |
0.2206853105 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 10 |
Hb_000130_330 |
0.2211008606 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 11 |
Hb_134949_010 |
0.2227906684 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 12 |
Hb_000175_640 |
0.2255367436 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_007803_040 |
0.2257621378 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000340_270 |
0.2283078411 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 15 |
Hb_026437_020 |
0.2318645962 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_001584_050 |
0.2318888692 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004611_040 |
0.2319717234 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001907_080 |
0.2320987731 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 19 |
Hb_000193_220 |
0.2322569977 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000107_540 |
0.2323717131 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |