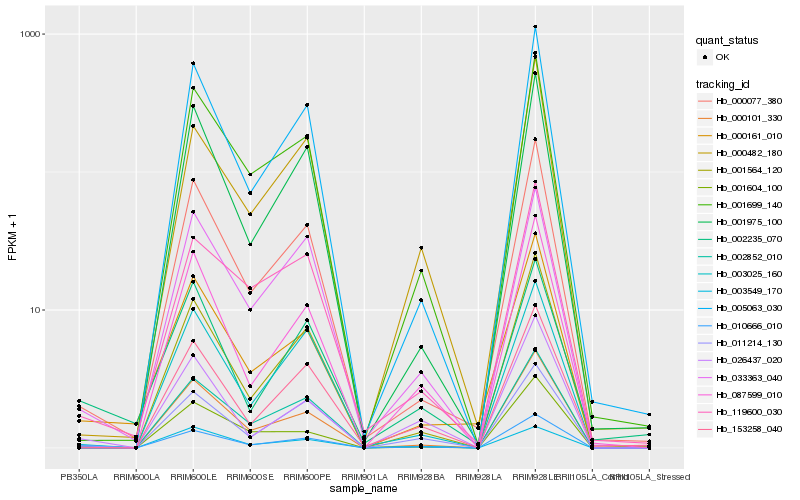
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010666_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000077_380 |
0.1239800588 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 3 |
Hb_011214_130 |
0.1251865078 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 4 |
Hb_000161_010 |
0.127917854 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_026437_020 |
0.1317159576 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_001564_120 |
0.1411286319 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 7 |
Hb_002235_070 |
0.1416080239 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 8 |
Hb_000101_330 |
0.1426123099 |
- |
- |
- |
| 9 |
Hb_033363_040 |
0.1426348472 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 10 |
Hb_005063_030 |
0.144554212 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_119600_030 |
0.1453993193 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 12 |
Hb_003549_170 |
0.1454196143 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 13 |
Hb_153258_040 |
0.1464159872 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 14 |
Hb_003025_160 |
0.1464200816 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 15 |
Hb_002852_010 |
0.1477223224 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_001699_140 |
0.1479276329 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 17 |
Hb_087599_010 |
0.1492156479 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 18 |
Hb_000482_180 |
0.1504788408 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 19 |
Hb_001604_100 |
0.1505621687 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 20 |
Hb_001975_100 |
0.1507534959 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |