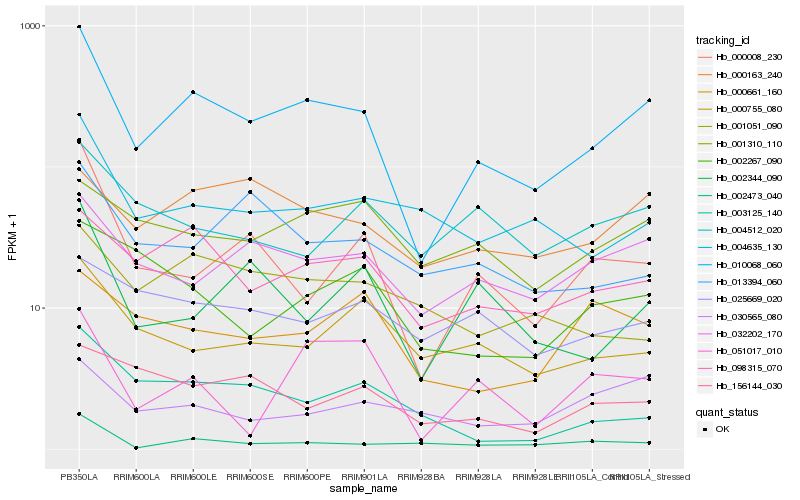
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010068_060 |
0.0 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 2 |
Hb_098315_070 |
0.1734986903 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 3 |
Hb_032202_170 |
0.1864336195 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 4 |
Hb_004635_130 |
0.201837329 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 5 |
Hb_000163_240 |
0.2053829406 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |
| 6 |
Hb_000008_230 |
0.2056790701 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 7 |
Hb_002344_090 |
0.2134549911 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 8 |
Hb_001310_110 |
0.21515011 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_002473_040 |
0.2154236753 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000755_080 |
0.215556916 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 11 |
Hb_002267_090 |
0.2205897444 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 12 |
Hb_013394_060 |
0.2215379491 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_004512_020 |
0.2245717315 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 14 |
Hb_025669_020 |
0.2249720923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_156144_030 |
0.2251168617 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 16 |
Hb_003125_140 |
0.2254731809 |
- |
- |
PREDICTED: uncharacterized protein LOC105648796 [Jatropha curcas] |
| 17 |
Hb_051017_010 |
0.225483782 |
- |
- |
Uncharacterized protein TCM_031047 [Theobroma cacao] |
| 18 |
Hb_001051_090 |
0.2257041093 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 19 |
Hb_030565_080 |
0.2259954978 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 20 |
Hb_000661_160 |
0.2283855292 |
- |
- |
hexokinase [Manihot esculenta] |