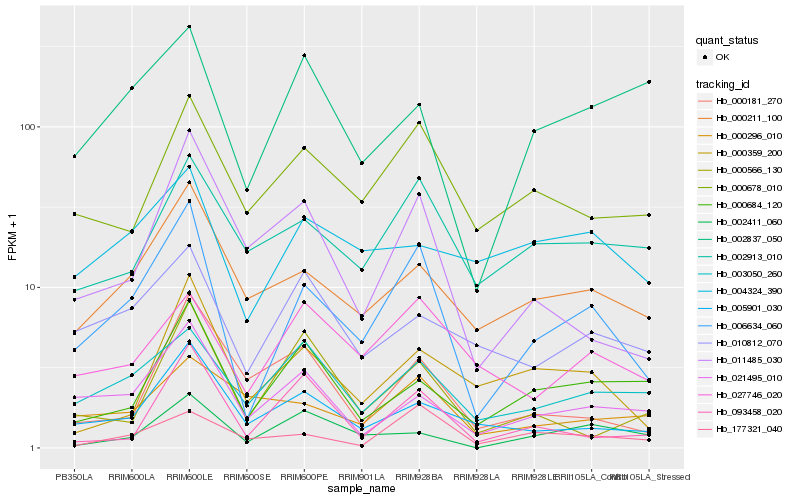
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006634_060 |
0.0 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_000211_100 |
0.2101919374 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 3 |
Hb_005901_030 |
0.2170634664 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_021495_010 |
0.2214644233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000678_010 |
0.2245943528 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_011485_030 |
0.227913099 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 7 |
Hb_003050_260 |
0.2303024742 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 8 |
Hb_002913_010 |
0.2343465746 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_027746_020 |
0.2412696287 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 10 |
Hb_177321_040 |
0.2413196674 |
- |
- |
hypothetical protein JCGZ_06328 [Jatropha curcas] |
| 11 |
Hb_000359_200 |
0.241552947 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_000684_120 |
0.2436537573 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010812_070 |
0.2464588335 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 14 |
Hb_093458_020 |
0.2512457583 |
desease resistance |
Gene Name: AAA |
PREDICTED: chromosome transmission fidelity protein 18 homolog [Jatropha curcas] |
| 15 |
Hb_000296_010 |
0.2555034817 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 16 |
Hb_002411_060 |
0.2570323188 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 17 |
Hb_002837_050 |
0.2587286203 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 18 |
Hb_004324_390 |
0.2589394102 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 19 |
Hb_000181_270 |
0.259909094 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000566_130 |
0.2599637325 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |